7VFM
 
 | | Crystal structure of SdgB (UDP and SD peptide-binding form) | | Descriptor: | Glycosyl transferase, group 1 family protein, SER-ASP-SER-ASP, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFN
 
 | | Crystal structure of SdgB (SD peptide-binding form) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFK
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4RNZ
 
 | | Structure of Helicobacter pylori Csd3 from the hexagonal crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RNY
 
 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JJT
 
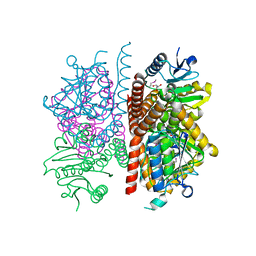 | | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv | | Descriptor: | ACETATE ION, Enoyl-CoA hydratase, GLYCEROL | | Authors: | Tan, K, Holowicki, J, Endres, M, Kim, C.-Y, Kim, H, Hung, L.-W, Terwilliger, T.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
2QHU
 
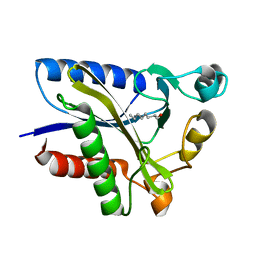 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTANAL | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2QHS
 
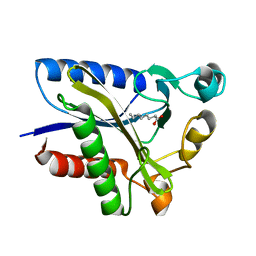 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2QHV
 
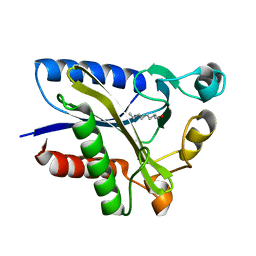 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTAN-1-OL | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2QHT
 
 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
6P5W
 
 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6P5V
 
 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
3MYP
 
 | | Crystal structure of tagatose-1,6-bisphosphate aldolase from Staphylococcus aureus | | Descriptor: | Tagatose 1,6-diphosphate aldolase | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Suh, S.W. | | Deposit date: | 2010-05-10 | | Release date: | 2011-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation
Febs Lett., 585, 2011
|
|
3MYO
 
 | | Crystal structure of tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes | | Descriptor: | Tagatose 1,6-diphosphate aldolase 1 | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Suh, S.W. | | Deposit date: | 2010-05-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation
Febs Lett., 585, 2011
|
|
5F4B
 
 | | Structure of B. abortus WrbA-related protein A (WrpA) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Herrou, J, Czyz, D, Willett, J.W, Kim, H.S, Chhor, G, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Crosson, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | WrpA Is an Atypical Flavodoxin Family Protein under Regulatory Control of the Brucella abortus General Stress Response System.
J.Bacteriol., 198, 2016
|
|
5F51
 
 | | Structure of B. abortus WrbA-related protein A (apo) | | Descriptor: | NAD(P)H dehydrogenase (quinone), SULFATE ION | | Authors: | Herrou, J, Czyz, D, Willett, J.W, Kim, H.S, Kim, Y, Crosson, S. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | WrpA Is an Atypical Flavodoxin Family Protein under Regulatory Control of the Brucella abortus General Stress Response System.
J.Bacteriol., 198, 2016
|
|
6OI6
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfide soaked) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, mitochondrial, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
3DCM
 
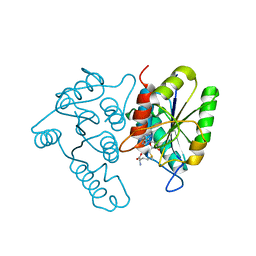 | | Crystal structure of the Thermotoga maritima SPOUT family RNA-methyltransferase protein Tm1570 in complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein TM_1570 | | Authors: | Kim, D.J, Kim, H.S, Lee, S.J, Suh, S.W. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thermotoga maritima SPOUT superfamily RNA methyltransferase Tm1570 in complex with S-adenosyl-L-methionine
Proteins, 74, 2009
|
|
6OIB
 
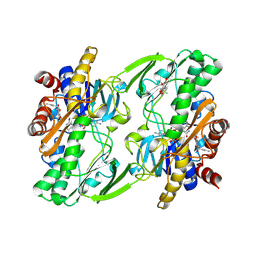 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
6OIC
 
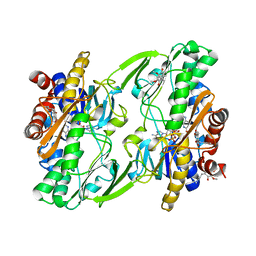 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfite soaked) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, Sulfide:quinone oxidoreductase, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
7WWP
 
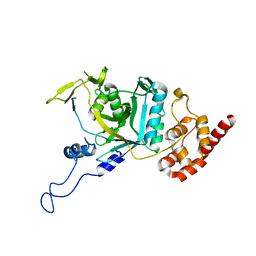 | | Crystal structure of human Npl4 | | Descriptor: | Nuclear protein localization protein 4 homolog, ZINC ION | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
7WWQ
 
 | | Crystal structure of human Ufd1-Npl4 complex | | Descriptor: | Nuclear protein localization protein 4 homolog, Ubiquitin recognition factor in ER-associated degradation protein 1 | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
1A77
 
 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1A76
 
 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
4XYH
 
 | |
