1L1J
 
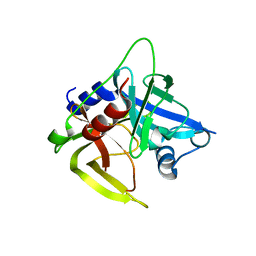 | | Crystal structure of the protease domain of an ATP-independent heat shock protease HtrA | | Descriptor: | heat shock protease HtrA | | Authors: | Kim, D.Y, Kim, D.R, Ha, S.C, Lokanath, N.K, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2002-02-18 | | Release date: | 2003-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Protease Domain of a Heat-shock Protein HtrA from Thermotoga maritima
J.BIOL.CHEM., 278, 2003
|
|
2ZL0
 
 | | Crystal structure of H.pylori ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
2ZL4
 
 | |
2ZL2
 
 | | Crystal structure of H.pylori ClpP in complex with the peptide NVLGFTQ | | Descriptor: | A peptide substrate-NVLGFTQ, A peptide substrate-NVLGFTQ for Chain R and S, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
2P4B
 
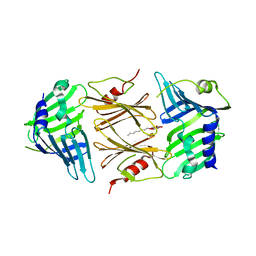 | | Crystal structure of E.coli RseB | | Descriptor: | Sigma-E factor regulatory protein rseB, octyl beta-D-glucopyranoside | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2007-03-12 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of RseB and a model of its binding mode to RseA
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ZL3
 
 | | Crystal structure of H.pylori ClpP S99A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
3M4W
 
 | | Structural basis for the negative regulation of bacterial stress response by RseB | | Descriptor: | Sigma-E factor negative regulatory protein, Sigma-E factor regulatory protein rseB, ZINC ION | | Authors: | Kim, D.Y, Kwon, E, Choi, J.K, Hwang, H.-Y, Kim, K.K. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the negative regulation of bacterial stress response by RseB
Protein Sci., 19, 2010
|
|
1UM8
 
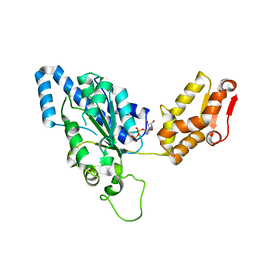 | | Crystal structure of helicobacter pylori ClpX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2003-09-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of ClpX Molecular Chaperone from Helicobacter pylori
J.Biol.Chem., 278, 2003
|
|
9KJB
 
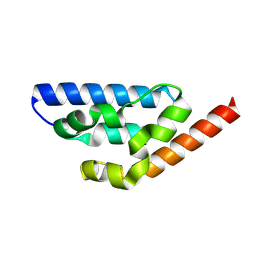 | | Crystal structure of Anti-CRISPR AcrIE7 | | Descriptor: | AcrIE7 | | Authors: | Kim, D.Y, Park, H.H. | | Deposit date: | 2024-11-12 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | AcrIE7 inhibits the CRISPR-Cas system by directly binding to the R-loop single-stranded DNA.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZEY
 
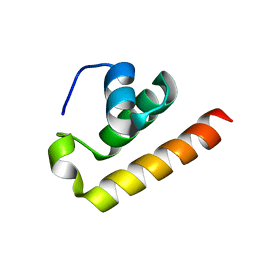 | | Anti-CRISPR type I subtype E3;AcrIE3 | | Descriptor: | AcrIE3 | | Authors: | Kim, D.Y, Park, H.H. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Novel structure of the anti-CRISPR protein AcrIE3 and its implication on the CRISPR-Cas inhibition.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
8ZNS
 
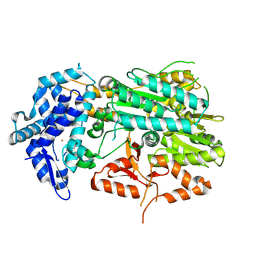 | | Type I-C CRISPR-associated protein, Cas3 | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, FE (III) ION | | Authors: | Kim, D.Y, Park, H.H. | | Deposit date: | 2024-05-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of Cas3 activation in type I-C CRISPR-Cas system.
Nucleic Acids Res., 52, 2024
|
|
4NJQ
 
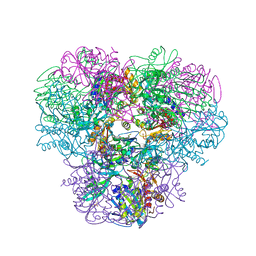 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CARBONATE ION, COBALT (II) ION, ... | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3BG4
 
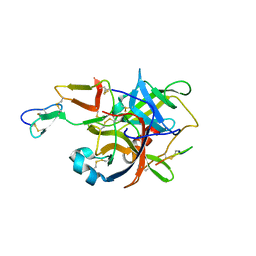 | | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Kim, H, Chu, T.T.T, Kim, D.Y, Kim, D.R, Nguyen, C.M.T, Choi, J, Lee, J.R, Hahn, M.J, Kim, K.K. | | Deposit date: | 2007-11-26 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor.
J.Mol.Biol., 376, 2008
|
|
4NJR
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3V67
 
 | | Periplasmic domain of Vibrio parahaemolyticus CpxA | | Descriptor: | Sensor protein CpxA | | Authors: | Kwon, E, Kim, D.Y, Ngo, T.D, Gross, J.D, Kim, K.K. | | Deposit date: | 2011-12-19 | | Release date: | 2012-09-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the periplasmic domain of Vibrio parahaemolyticus CpxA
Protein Sci., 21, 2012
|
|
5X55
 
 | |
1YGZ
 
 | | Crystal Structure of Inorganic Pyrophosphatase from Helicobacter pylori | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Wu, C.A, Lokanath, N.K, Kim, D.Y, Park, H.J, Hwang, H.Y, Kim, S.T, Suh, S.W, Kim, K.K. | | Deposit date: | 2005-01-06 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of inorganic pyrophosphatase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2P52
 
 | |
3OEO
 
 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
7WA4
 
 | | Crystal structure of GIGANTEA in complex with LKP2 | | Descriptor: | Adagio protein 2, FLAVIN MONONUCLEOTIDE, Protein GIGANTEA | | Authors: | Pathak, D, Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the regulation of blue-light receptors by GIGANTEA.
Cell Rep, 39, 2022
|
|
6LYE
 
 | | Crystal Structure of mimivirus UNG Y322F in complex with UGI | | Descriptor: | Probable uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
J.Struct.Biol., 211, 2020
|
|
5WUQ
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, a zinc binding form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW, ZINC ION | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5WUR
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, an oxdized form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
8I2E
 
 | |
8I2F
 
 | |
