1BJ8
 
 | | THIRD N-TERMINAL DOMAIN OF GP130, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GP130 | | Authors: | Kernebeck, T, Pflanz, S, Muller-Newen, G, Kurapkat, G, Scheek, R.M, Dijkstra, K, Heinrich, P.C, Wollmer, A, Grzesiek, S, Grotzinger, J. | | Deposit date: | 1998-07-02 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The signal transducer gp130: solution structure of the carboxy-terminal domain of the cytokine receptor homology region.
Protein Sci., 8, 1999
|
|
5NPA
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
2M09
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
4BA8
 
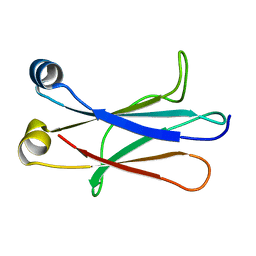 | | High resolution NMR structure of the C mu3 domain from IgM | | Descriptor: | IG MU CHAIN C REGION SECRETED FORM | | Authors: | Mueller, R, Kern, T, Graewert, M.A, Madl, T, Peschek, J, Groll, M, Sattler, M, Buchner, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-06-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High Resolution Structures of the Igm Fc Domains Reveal Principles of its Hexamer Formation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JVU
 
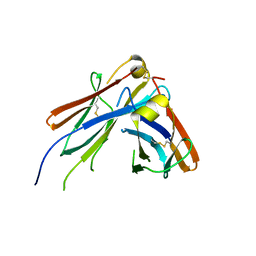 | | IgM C2-domain from mouse | | Descriptor: | Ig mu chain C region membrane-bound form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JVW
 
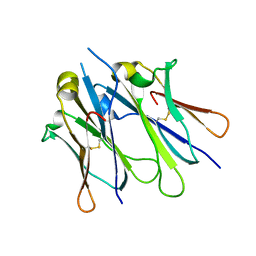 | | IgM C4-domain from mouse | | Descriptor: | Ig mu chain C region secreted form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QMY
 
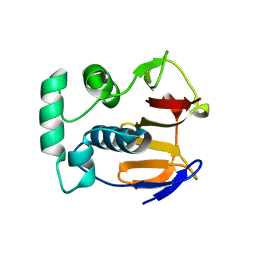 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
7ARA
 
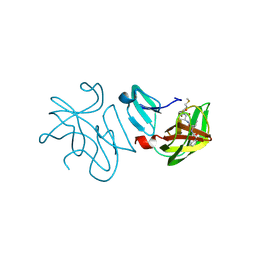 | | Rhinovirus A2 2A protease in complex with zVAM.fmk | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-3-methyl-1-[[(2~{S})-1-[[(3~{S})-1-methylsulfanyl-4-oxidanylidene-pentan-3-yl]amino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, (phenylmethyl) ~{N}-[(2~{S})-3-methyl-1-[[(2~{R})-1-[[(3~{R})-1-methylsulfanyl-4-oxidanylidene-pentan-3-yl]amino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 2A protease, ... | | Authors: | Deutschmann-Olek, K.M, Skern, T, Bezerra, G.A, Yue, W.W. | | Deposit date: | 2020-10-23 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Defining substrate selection by rhinoviral 2A proteinase through its crystal structure with the inhibitor zVAM.fmk.
Virology, 562, 2021
|
|
5EZU
 
 | | Crystal structure of the N-terminal domain of vaccinia virus immunomodulator A46 in complex with myristic acid. | | Descriptor: | MYRISTIC ACID, Protein A46 | | Authors: | Fedosyuk, S, Bezerra, G.A, Sammito, M, Uson, I, Skern, T. | | Deposit date: | 2015-11-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog., 12, 2016
|
|
6S92
 
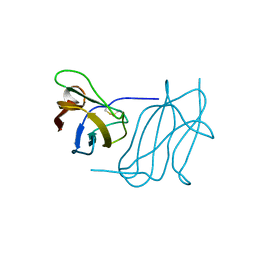 | |
6S94
 
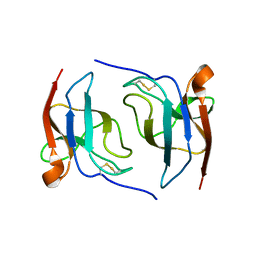 | |
6S95
 
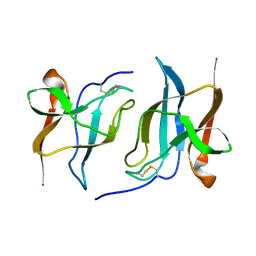 | |
6S93
 
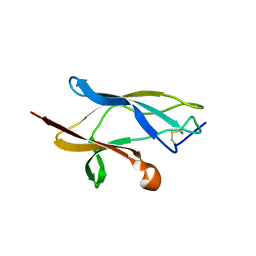 | |
4QBB
 
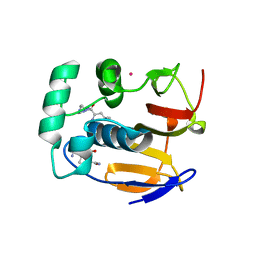 | | Structure of the foot-and-mouth disease virus leader proteinase in complex with inhibitor (N~2~-[(3S)-4-({(2R)-1-[(4-CARBAMIMIDAMIDOBUTYL)AMINO]-4-METHYL-1-OXOPENTAN-2-YL}AMINO)-3-HYDROXY-4-OXOBUTANOYL]-L-ARGINYL-L-PROLINAMIDE) | | Descriptor: | Leader protease, N~2~-[(3S)-4-({(2R)-1-[(4-carbamimidamidobutyl)amino]-4-methyl-1-oxopentan-2-yl}amino)-3-hydroxy-4-oxobutanoyl]-L-arginyl-L-prolinamide, PHOSPHATE ION, ... | | Authors: | Grishkovskaya, I, Steinberger, J, Cencic, R, Juliano, M.A, Juliano, L, Skern, T. | | Deposit date: | 2014-05-07 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Foot-and-mouth disease virus leader proteinase: Structural insights into the mechanism of intermolecular cleavage.
Virology, 468-470C, 2014
|
|
1Z8R
 
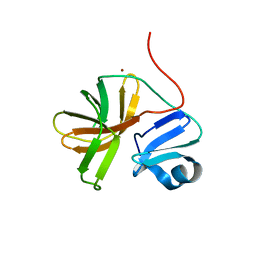 | | 2A cysteine proteinase from human coxsackievirus B4 (strain JVB / Benschoten / New York / 51) | | Descriptor: | Coxsackievirus B4 polyprotein, ZINC ION | | Authors: | Baxter, N.J, Roetzer, A, Liebig, H.D, Sedelnikova, S.E, Hounslow, A.M, Skern, T, Waltho, J.P. | | Deposit date: | 2005-03-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of coxsackievirus B4 2A proteinase, an enyzme involved in the etiology of heart disease.
J.Virol., 80, 2006
|
|
2JQF
 
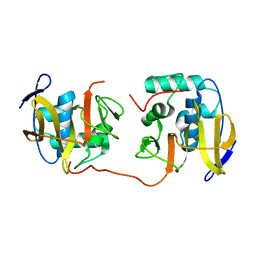 | | Full Length Leader Protease of Foot and Mouth Disease Virus C51A Mutant | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
2HRV
 
 | | 2A CYSTEINE PROTEINASE FROM HUMAN RHINOVIRUS 2 | | Descriptor: | 2A CYSTEINE PROTEINASE, ZINC ION | | Authors: | Petersen, J.F.W, Cherney, M.M, Liebig, H.-D, Skern, T, Kuechler, E, James, M.N.G. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis.
EMBO J., 18, 1999
|
|
1QOL
 
 | | STRUCTURE OF THE FMDV LEADER PROTEASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROTEASE (NONSTRUCTURAL PROTEIN P20A) | | Authors: | Guarne, A, Tormo, J, Kirchweger, R, Pfistermueller, D, Skern, T, Fita, I. | | Deposit date: | 1999-11-13 | | Release date: | 2000-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Foot-and-Mouth Disease Virus Leader Protease: A Papain-Like Fold Adapted for Self-Processing and Eif4G Recognition.
Embo J., 17, 1998
|
|
2JQG
 
 | | Leader Protease | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
4LQK
 
 | | Structure of the vaccinia virus NF- B antagonist A46 | | Descriptor: | BROMIDE ION, Protein A46, SODIUM ION | | Authors: | Grishkovskaya, I, Fedosyuk, S, Skern, T, Djinovic-Carugo, K. | | Deposit date: | 2013-07-18 | | Release date: | 2013-12-25 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization and Structure of the Vaccinia Virus NF-kappa B Antagonist A46.
J.Biol.Chem., 289, 2014
|
|
