3GI0
 
 | |
2LET
 
 | | AN 1H NMR DETERMINATION OF THE THREE DIMENSIONAL STRUCTURES OF MIRROR IMAGE FORMS OF A LEU-5 VARIANT OF THE TRYPSIN INHIBITOR ECBALLIUM ELATERIUM (EETI-II) | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Nielsen, K.J, Alewood, D, Andrews, J, Kent, S.B.H, Craik, D.J. | | Deposit date: | 1994-01-04 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | An 1H NMR determination of the three-dimensional structures of mirror-image forms of a Leu-5 variant of the trypsin inhibitor from Ecballium elaterium (EETI-II).
Protein Sci., 3, 1994
|
|
3E8Y
 
 | | Xray structure of scorpion toxin BmBKTx1 | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 19.1, SULFATE ION | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-20 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structure of native scorpion toxin BmBKTx1 by racemic protein crystallography using direct methods.
J.Am.Chem.Soc., 131, 2009
|
|
3BOG
 
 | | Snow Flea Antifreeze Protein Quasi-racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein, UNKNOWN LIGAND | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
2O40
 
 | |
3E7R
 
 | | X-ray Crystal Structure of Racemic Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3BOI
 
 | | Snow Flea Antifreeze Protein Racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
2NUI
 
 | | X-ray Structure of synthetic [D83A]RNase A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Kent, S.B.H. | | Deposit date: | 2006-11-09 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Total synthesis by modern chemical ligation methods and high resolution (1.1 A) X-ray structure of ribonuclease A
Biopolymers, 90, 2008
|
|
2NWD
 
 | |
3E7U
 
 | | X-ray Crystal Structure of L-Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3DCK
 
 | | X-ray structure of D25N chemical analogue of HIV-1 protease complexed with ketomethylene isostere inhibitor | | Descriptor: | (2S)-2-{[(2R,5S)-5-{[(2S,3S)-2-{[(2S,3R)-2-(acetylamino)-3-hydroxybutanoyl]amino}-3-methylpentanoyl]amino}-2-butyl-4-oxononanoyl]amino}-N~1~-[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]pentanediamide, Chemical analogue HIV-1 protease | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DCR
 
 | | X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor | | Descriptor: | Chemical analogue HIV-1 protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2OQF
 
 | | Structure of a synthetic, non-natural analogue of RNase A: [N71K(Ade), D83A]RNase A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Zhang, J.L, He, C, Kent, S.B.H. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Characterization of Non-natural RNase A Analogues with Enhanced Second-step Catalytic Activity
To be Published
|
|
2FD7
 
 | | X-ray Crystal Structure of Chemically Synthesized Crambin | | Descriptor: | Crambin | | Authors: | Bang, D, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Mol Biosyst, 5, 2009
|
|
2FCQ
 
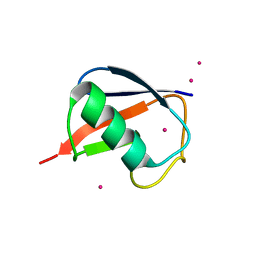 | | X-ray Crystal Structure of a Chemically Synthesized Ubiquitin with a Cubic Space Group | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCM
 
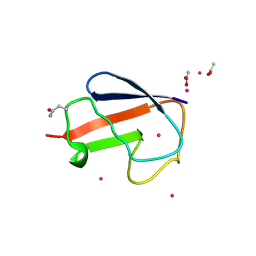 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCS
 
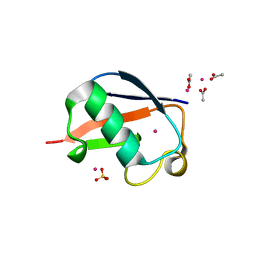 | | X-ray Crystal Structure of a Chemically Synthesized [L-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, SULFATE ION, ... | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCN
 
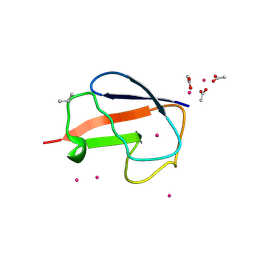 | | X-ray Crystal Structure of a Chemically Synthesized [D-Val35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FD9
 
 | | X-ray Crystal Structure of Chemically Synthesized Crambin-{alpha}carboxamide | | Descriptor: | Crambin | | Authors: | Bang, D, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Mol Biosyst, 5, 2009
|
|
