6D0Q
 
 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.80051017 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
6D0R
 
 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85856962 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
2OUA
 
 | | Crystal Structure of Nocardiopsis Protease (NAPase) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic exploration of Acid resistance: kinetic stability facilitates evolution of extremophilic behavior
J.Mol.Biol., 368, 2007
|
|
2PFE
 
 | | Crystal Structure of Thermobifida fusca Protease A (TFPA) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Alkaline serine protease, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.436 Å) | | Cite: | Mesophile versus Thermophile: Insights Into the Structural Mechanisms of Kinetic Stability
J.Mol.Biol., 370, 2007
|
|
3URD
 
 | | T181A mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3URC
 
 | | T181G mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3URE
 
 | |
3U5Z
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3U60
 
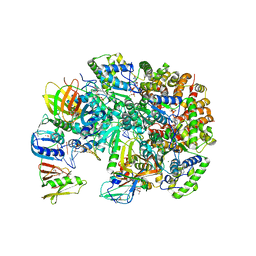 | | Structure of T4 Bacteriophage Clamp Loader Bound To Open Clamp, DNA and ATP Analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3U61
 
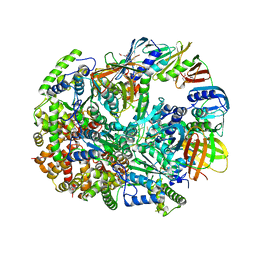 | | Structure of T4 Bacteriophage Clamp Loader Bound To Closed Clamp, DNA and ATP Analog and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
8ED0
 
 | |
8EDX
 
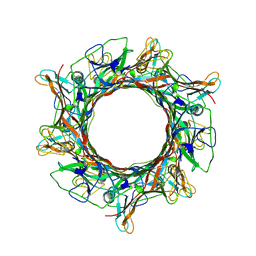 | |
6O3H
 
 | | Icosahedral reconstruction of the thermophilic bacteriophage P74-26 capsid | | Descriptor: | Major head protein, P74-26 Head Decoration Protein | | Authors: | Stone, N.P, Demo, G, Agnello, E, Kelch, B.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure.
Nat Commun, 10, 2019
|
|
5TGE
 
 | |
7U19
 
 | | RFC:PCNA bound to nicked DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA, MAGNESIUM ION, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7U1P
 
 | | RFC:PCNA bound to DNA with a ssDNA gap of five nucleotides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
8E84
 
 | | Human PCNA mutant- C148S | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Magrino, J, Page, B, Gaubitz, C, Kelch, B.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A thermosensitive PCNA allele underlies an ataxia-telangiectasia-like disorder.
J.Biol.Chem., 299, 2023
|
|
7U1A
 
 | | RFC:PCNA bound to dsDNA with a ssDNA gap of six nucleotides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
