6ZWV
 
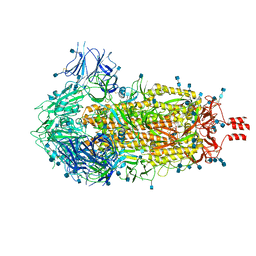 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
7OVR
 
 | | Mature HIV-1 matrix structure | | Descriptor: | HIV-1 matrix, MYRISTIC ACID, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
7OVQ
 
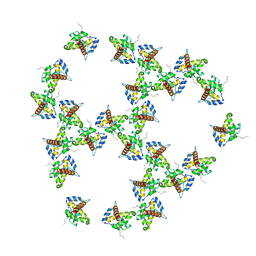 | | Immature HIV-1 matrix structure | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
3RHY
 
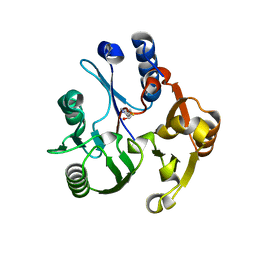 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with 4-chloro-2-hydroxymethylpyridine | | Descriptor: | (4-chloropyridin-2-yl)methanol, N(G),N(G)-dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Johnson, C.M, Ke, Z, Yoon, D.-W, Linsky, T.W, Guo, H, Fast, W, Robertus, J.D. | | Deposit date: | 2011-04-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | On the mechanism of dimethylarginine dimethylaminohydrolase inactivation by 4-halopyridines.
J.Am.Chem.Soc., 133, 2011
|
|
6XL0
 
 | | Caulobacter crescentus FljK filament | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
6XKY
 
 | | Caulobacter crescentus FljK filament, straightened | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
5YNO
 
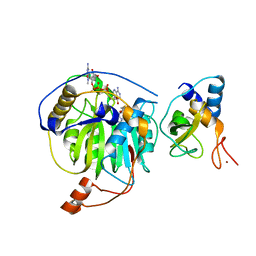 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
8XAB
 
 | |
8X0A
 
 | |
5YN8
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNJ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN6
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex bound to SAM | | Descriptor: | S-ADENOSYLMETHIONINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNN
 
 | | Crystal structure of MERS-CoV nsp16/nsp10complex bound to sinefungin and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNI
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNQ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN5
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNF
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNP
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNB
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to Sinefungin | | Descriptor: | SINEFUNGIN, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNM
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
7EQ4
 
 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
