7Y9Y
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | 分子名称: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | 著者 | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-06-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
 | | Structure of the Cas7-11-Csx29-guide RNA complex | | 分子名称: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | 著者 | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-06-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
5X32
 
 | | Crystal structure of D-mannose isomerase | | 分子名称: | N-acylglucosamine 2-epimerase, PHOSPHATE ION | | 著者 | Kato, K, Saburi, W, Yao, M. | | 登録日 | 2017-02-03 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.586 Å) | | 主引用文献 | Biochemical and structural characterization of Marinomonas mediterranead-mannose isomerase Marme_2490 phylogenetically distant from known enzymes
Biochimie, 144, 2018
|
|
7DR0
 
 | | Structure of Wild-type PSI monomer1 from Cyanophora paradoxa | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | 登録日 | 2020-12-25 | | 公開日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7DR2
 
 | | Structure of GraFix PSI tetramer from Cyanophora paradoxa | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | 登録日 | 2020-12-25 | | 公開日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7DR1
 
 | | Structure of Wild-type PSI monomer2 from Cyanophora paradoxa | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | 登録日 | 2020-12-25 | | 公開日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
5ZCC
 
 | |
5ZCE
 
 | | Crystal structure of Alpha-glucosidase in complex with maltotetraose | | 分子名称: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Kato, K, Saburi, W, Yao, M. | | 登録日 | 2018-02-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.555 Å) | | 主引用文献 | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5ZCD
 
 | | Crystal structure of Alpha-glucosidase in complex with maltotriose | | 分子名称: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Kato, K, Saburi, W, Yao, M. | | 登録日 | 2018-02-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.707 Å) | | 主引用文献 | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5ZCB
 
 | | Crystal structure of Alpha-glucosidase | | 分子名称: | Alpha-glucosidase, CALCIUM ION | | 著者 | Kato, K, Saburi, W, Yao, M. | | 登録日 | 2018-02-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
6AEK
 
 | | Crystal structure of ENPP1 in complex with pApG | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-MONOPHOSPHATE, ... | | 著者 | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-08-05 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
7JL3
 
 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | 著者 | Kato, K, Ahmad, S, Hur, S. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL0
 
 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | 著者 | Kato, K, Ahmad, S, Hur, S. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL1
 
 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | 著者 | Kato, K, Ahmad, S, Hur, S. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL4
 
 | | Crystal structure of TRIM65 PSpry domain | | 分子名称: | GLYCEROL, Tripartite motif-containing protein 65 | | 著者 | Kato, K, Ahmad, S, Hur, S. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL2
 
 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | 著者 | Kato, K, Ahmad, S, Hur, S. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
6AEL
 
 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | 著者 | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-08-05 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
7WAH
 
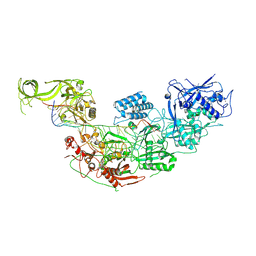 | | Structure of Cas7-11 in complex with guide RNA and target RNA | | 分子名称: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (39-MER), ... | | 著者 | Kato, K, Okazaki, S, Isayama, Y, Nishizawa, T, Nishimasu, H. | | 登録日 | 2021-12-14 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex.
Cell, 185, 2022
|
|
7XHT
 
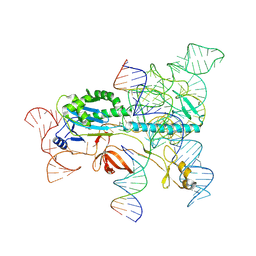 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | 分子名称: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-04-10 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
2DJJ
 
 | |
2DJK
 
 | |
3WT0
 
 | |
7VQP
 
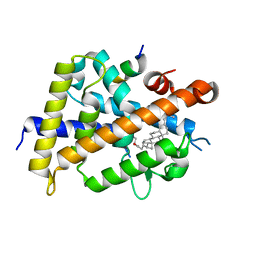 | | Vitamin D receptor complexed with a lithocholic acid derivative | | 分子名称: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | 登録日 | 2021-10-20 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
3J7T
 
 | | Calcium atpase structure with two bound calcium ions determined by electron crystallography of thin 3D crystals | | 分子名称: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | 著者 | Yonekura, K, Kato, K, Ogasawara, M, Tomita, M, Toyoshima, C. | | 登録日 | 2014-08-07 | | 公開日 | 2015-02-18 | | 最終更新日 | 2016-09-28 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.4 Å) | | 主引用文献 | Electron crystallography of ultrathin 3D protein crystals: atomic model with charges
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1NPM
 
 | | NEUROPSIN, A SERINE PROTEASE EXPRESSED IN THE LIMBIC SYSTEM OF MOUSE BRAIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUROPSIN | | 著者 | Kishi, T, Kato, M, Shimizu, T, Kato, K, Matsumoto, K, Yoshida, S, Shiosaka, S, Hakoshima, T. | | 登録日 | 1998-01-07 | | 公開日 | 1999-03-23 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis.
J.Biol.Chem., 274, 1999
|
|
