8PE8
 
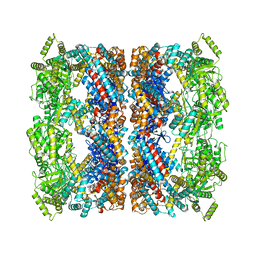 | |
8PE4
 
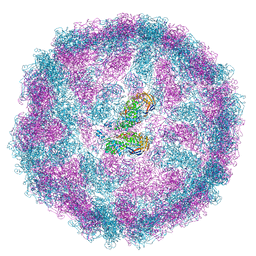 | |
8OIU
 
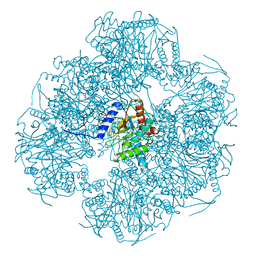 | | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Skalidis, I, Tueting, C, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon.
Commun Biol, 6, 2023
|
|
9F2K
 
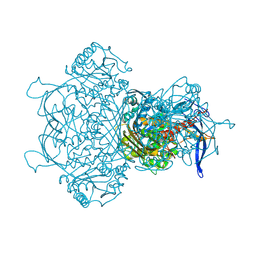 | | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, inositol-3-phosphate synthase | | Authors: | Traeger, T.K, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Disorder-to-order active site capping regulates the rate-limiting step of the inositol pathway.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9IG6
 
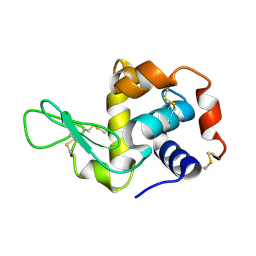 | | Hen egg-white lysozyme structure determined by 3DED/MicroED on a 200 keV microscope | | Descriptor: | Lysozyme C | | Authors: | Shaikhqasem, A, Hamdi, F, Machner, L, Parthier, C, Breithaupt, C, Kyrilis, F.L, Feller, S.M, Kastritis, P.L, Stubbs, M.T. | | Deposit date: | 2025-02-19 | | Release date: | 2025-03-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Hen egg-white lysozyme structure determined by 3DED/MicroED on a 200 keV microscope
To Be Published
|
|
7OTT
 
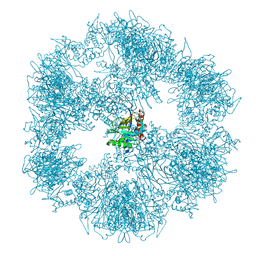 | |
7BGJ
 
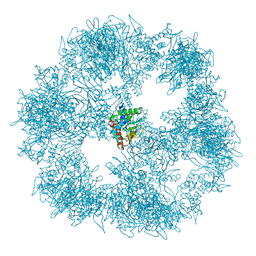 | | C. thermophilum Pyruvate Dehydrogenase Complex Core | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Tueting, C, Kastritis, P.L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts.
Cell Rep, 34, 2021
|
|
6SHT
 
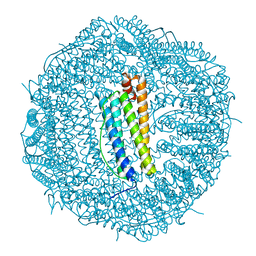 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
8A5T
 
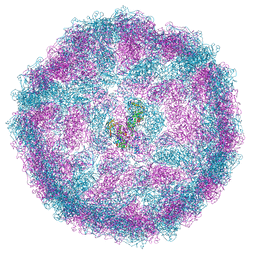 | |
7Q5R
 
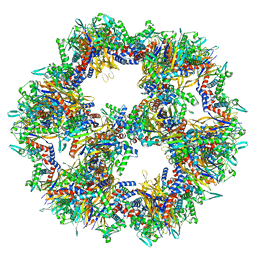 | | Protein community member pyruvate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5Q
 
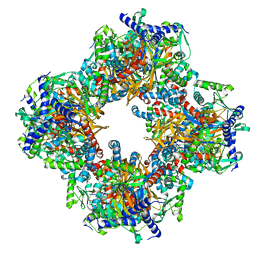 | | Protein community member oxoglutarate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5S
 
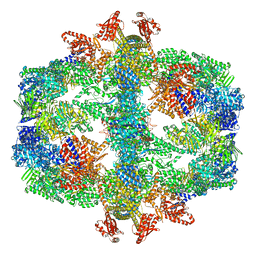 | | Protein community member fatty acid synthase complex from C. thermophilum | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
5UCY
 
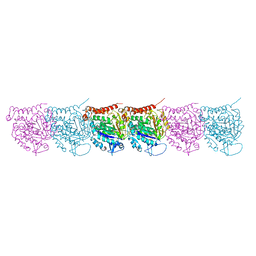 | | Cryo-EM map of protofilament of microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
5UBQ
 
 | | Cryo-EM structure of ciliary microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
8S5C
 
 | | Cryo-EM structure of Arf1-decorated membrane tubules | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosylation factor 1, MAGNESIUM ION | | Authors: | Haupt, C, Semchonok, D.A, Stubbs, M.T, Bacia, K, Desfosses, A, Kastritis, P.L, Hamdi, F. | | Deposit date: | 2024-02-23 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Arf1-decorated membrane tubules
To Be Published
|
|
8S5D
 
 | | Cryo-EM structure of Arf1-decorated membrane tubules | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosylation factor 1, MAGNESIUM ION | | Authors: | Haupt, C, Semchonok, D.A, Stubbs, M.T, Bacia, K, Desfosses, A, Kastritis, P.L, Hamdi, F. | | Deposit date: | 2024-02-23 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structure of Arf1-decorated membrane tubules
To Be Published
|
|
8S5E
 
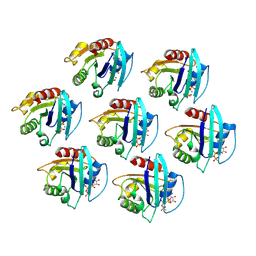 | | Cryo-EM structure of Arf1-decorated membrane tubules | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosylation factor 1, MAGNESIUM ION | | Authors: | Haupt, C, Semchonok, D.A, Stubbs, M.T, Bacia, K, Desfosses, A, Kastritis, P.L, Hamdi, F. | | Deposit date: | 2024-02-23 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Arf1-decorated membrane tubules
Cell Structure, 2024
|
|
