5X7K
 
 | |
1IS1
 
 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
1ISE
 
 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
2ZXC
 
 | | Ceramidase complexed with C2 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Okano, H, Inoue, T, Okino, N, Kakuta, Y, Matsumura, H, Ito, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
3AHO
 
 | | PZ PEPTIDASE A with inhibitor 2 | | Descriptor: | 1-{3-[(R)-{(1R)-1-[(glycyl-L-prolyl)amino]-2-phenylethyl}(hydroxy)phosphoryl]propanoyl}-L-prolyl-D-norleucine, ACETATE ION, Oligopeptidase, ... | | Authors: | Nakano, H. | | Deposit date: | 2010-04-25 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
3AHN
 
 | | PZ PEPTIDASE A with Inhibitor 1 | | Descriptor: | ACETATE ION, N~2~-{(2S)-3-[(R)-hydroxy{(1R)-2-phenyl-1-[(phenylacetyl)amino]ethyl}phosphoryl]-2-methylpropanoyl}-L-lysyl-D-serine, Oligopeptidase, ... | | Authors: | Nakano, H. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
3AHM
 
 | | Pz peptidase a | | Descriptor: | ACETATE ION, Oligopeptidase, ZINC ION | | Authors: | Nakano, H. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
3X17
 
 | | Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase | | Descriptor: | CALCIUM ION, Endoglucanase, ZINC ION | | Authors: | Okano, H, Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, activity, and stability of metagenome-derived glycoside hydrolase family 9 endoglucanase with an N-terminal Ig-like domain.
Protein Sci., 24, 2015
|
|
3WX5
 
 | |
3WYD
 
 | |
1BJ3
 
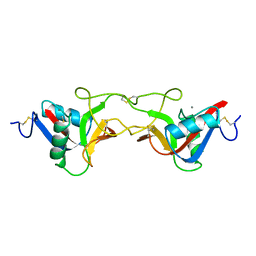 | | CRYSTAL STRUCTURE OF COAGULATION FACTOR IX-BINDING PROTEIN (IX-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | Descriptor: | CALCIUM ION, PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN A), PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN B) | | Authors: | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H, Atoda, H, Morita, T. | | Deposit date: | 1998-07-02 | | Release date: | 1999-08-16 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 A: implication of central loop swapping based on deletion in the linker region.
J.Mol.Biol., 289, 1999
|
|
1IXX
 
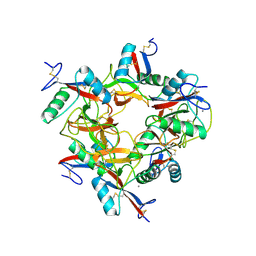 | | CRYSTAL STRUCTURE OF COAGULATION FACTORS IX/X-BINDING PROTEIN (IX/X-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | Descriptor: | CALCIUM ION, COAGULATION FACTORS IX/X-BINDING PROTEIN | | Authors: | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H. | | Deposit date: | 1997-05-01 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of coagulation factors IX/X-binding protein, a heterodimer of C-type lectin domains.
Nat.Struct.Biol., 4, 1997
|
|
5IB9
 
 | |
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
2D1X
 
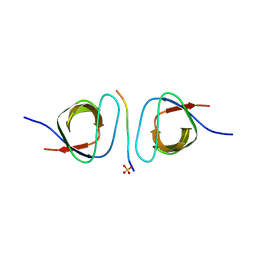 | | The crystal structure of the cortactin-SH3 domain and AMAP1-peptide complex | | Descriptor: | SULFATE ION, cortactin isoform a, proline rich region from development and differentiation enhancing factor 1 | | Authors: | Hashimoto, S, Hirose, M, Hashimoto, A, Morishige, M, Yamada, A, Hosaka, H, Akagi, K, Ogawa, E, Oneyama, C, Agatsuma, T, Okada, M, Kobayashi, H, Wada, H, Nakano, H, Ikegami, T, Nakagawa, A, Sabe, H. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2EK8
 
 | |
2EK9
 
 | |
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
1J0S
 
 | | Solution structure of the human interleukin-18 | | Descriptor: | Interleukin-18 | | Authors: | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
2DT4
 
 | | Crystal structure of Pyrococcus horikoshii a plant- and prokaryote-conserved (PPC) protein at 1.60 resolution | | Descriptor: | GLYCEROL, Hypothetical protein PH0802 | | Authors: | Lin, L, Nakano, H, Uchiyama, S, Fujimoto, S, Matsunaga, S, Nakamura, S. | | Deposit date: | 2006-07-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Pyrococcus horikoshii PPC protein at 1.60 A resolution
Proteins, 67, 2007
|
|
3TK5
 
 | | Factor Xa in complex with D102-4380 | | Descriptor: | 4-{3-[(4-chlorophenyl)amino]-3-oxopropyl}-3-({[5-(propan-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]carbonyl}amino)benzoic acid, CALCIUM ION, Factor X heavy chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
3TK6
 
 | | factor Xa in complex with D46-5241 | | Descriptor: | CALCIUM ION, Factor X heavy chain, Factor X light chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
2ZZO
 
 | | Crystal structure of the complex between GP41 fragment N36 and fusion inhibitor C34/S138A | | Descriptor: | Transmembrane protein | | Authors: | Watabe, T, Nakano, H, Nakatsu, T, Kato, H, Fujii, N. | | Deposit date: | 2009-02-20 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic study of an HIV-1 fusion inhibitor with the gp41 S138A substitution
J.Mol.Biol., 392, 2009
|
|
2ZWS
 
 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
