6UG0
 
 | | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2019-09-25 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
4M6R
 
 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | Descriptor: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | Authors: | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | Deposit date: | 2013-08-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7MCI
 
 | | MoFe protein from Azotobacter vinelandii with a sulfur-replenished cofactor | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Lee, C, Hu, Y, Ribbe, M.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nat Catal, 5, 2022
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
7JMA
 
 | |
8KG2
 
 | |
7JMB
 
 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | Authors: | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8K1H
 
 | |
8K1G
 
 | |
8HL7
 
 | |
8HL6
 
 | |
6NZJ
 
 | | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO2 Capture by a Surface-Exposed [Fe4S4] Cluster | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein, SULFATE ION | | Authors: | Rettberg, L.A, Kang, W, Stiebritz, M.T, Hiller, C.J, Lee, C.C, Liedtke, J, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO 2 Capture by a Surface-Exposed [Fe 4 S 4 ] Cluster.
Mbio, 10, 2019
|
|
3QC8
 
 | | Crystal Structure of FAF1 UBX Domain In Complex with p97/VCP N Domain Reveals The Conserved FcisP Touch-Turn Motif of UBX Domain Suffering Conformational Change | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Kim, K.H, Kang, W, Suh, S.W, Yang, J.K. | | Deposit date: | 2011-01-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain
Proteins, 79, 2011
|
|
6O0B
 
 | | Structural and Mechanistic Insights into CO2 Activation by Nitrogenase Iron Protein | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein | | Authors: | Rettberg, L.A, Stiebritz, M.T, Kang, W, Lee, C.C, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights into CO2Activation by Nitrogenase Iron Protein.
Chemistry, 25, 2019
|
|
7VID
 
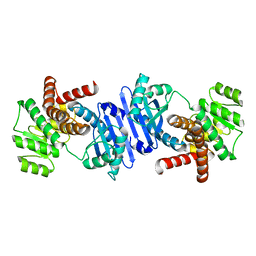 | |
3QCA
 
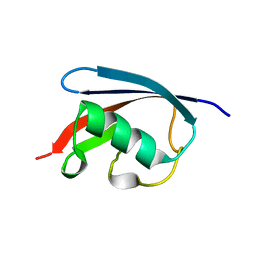 | |
7WWP
 
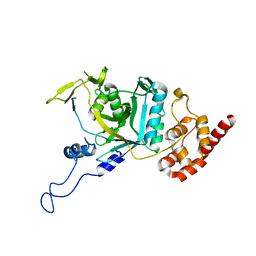 | | Crystal structure of human Npl4 | | Descriptor: | Nuclear protein localization protein 4 homolog, ZINC ION | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
7WWQ
 
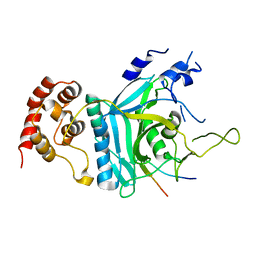 | | Crystal structure of human Ufd1-Npl4 complex | | Descriptor: | Nuclear protein localization protein 4 homolog, Ubiquitin recognition factor in ER-associated degradation protein 1 | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
8JGC
 
 | | Cryo-EM structure of Mi3 fused with LOV2 | | Descriptor: | LOV domain-containing protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8JGA
 
 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8HRZ
 
 | |
