6UCX
 
 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
4LBH
 
 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Apo-form | | Descriptor: | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG), DI(HYDROXYETHYL)ETHER | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4LBI
 
 | |
4KE4
 
 | | Elucidation of the structure and reaction mechanism of Sorghum bicolor hydroxycinnamoyltransferase and its structural relationship to other CoA-dependent transferases and synthases | | Descriptor: | GLYCEROL, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase, TETRAETHYLENE GLYCOL | | Authors: | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
4LTD
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - apo form | | Descriptor: | NADH-dependent FMN reductase, PHOSPHATE ION, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4LTN
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN, NADH complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, ... | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
1QYD
 
 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | pinoresinol-lariciresinol reductase | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
1QYC
 
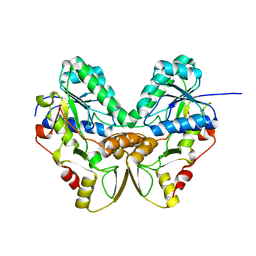 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | phenylcoumaran benzylic ether reductase PT1 | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
1RF4
 
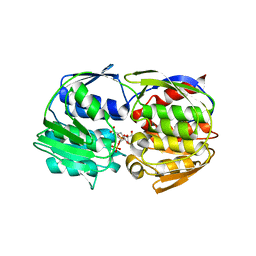 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1RH1
 
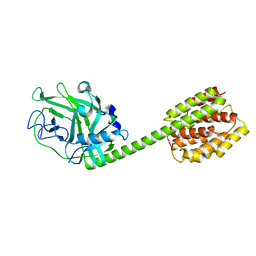 | | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | | Descriptor: | Colicin B | | Authors: | Hilsenbeck, J.L, Park, H, Chen, G, Youn, B, Postle, K, Kang, C. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution
Mol.Microbiol., 51, 2004
|
|
1RF5
 
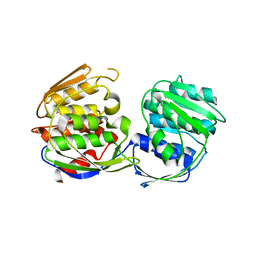 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in Unliganded State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1RF6
 
 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1SMM
 
 | | Crystal Structure of Cp Rd L41A mutant in oxidized state | | Descriptor: | FE (III) ION, Rubredoxin, SULFATE ION | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1SM5
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2004-03-08 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SMU
 
 | | Crystal Structure of Cp Rd L41A mutant in reduced state 1 (drop-reduced) | | Descriptor: | FE (II) ION, Rubredoxin | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1SMW
 
 | | Crystal Structure of Cp Rd L41A mutant in reduced state 2 (soaked) | | Descriptor: | FE (II) ION, Rubredoxin | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1T9P
 
 | | Crystal Structure of V44A, G45P Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1T4I
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.S, Kang, C. | | Deposit date: | 2004-04-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a cis-syn Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1T9O
 
 | | Crystal Structure of V44G Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
4YSB
 
 | | Crystal structure of ETHE1 from Myxococcus xanthus | | Descriptor: | FE (III) ION, Metallo-beta-lactamase family protein | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5015 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
4YSK
 
 | | Crystal structure of apo-form SdoA from Pseudomonas putida | | Descriptor: | Beta-lactamase domain protein, FE (III) ION | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.466 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
2LAT
 
 | | Solution structure of a Human minimembrane protein OST4 | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 4 | | Authors: | Gayen, S, Kang, C. | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human minimembrane protein Ost4, a subunit of the oligosaccharyltransferase complex.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
2M0S
 
 | |
2MFR
 
 | |
