2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
6AFK
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
2J3H
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | Descriptor: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
2W9Q
 
 | | Crystal Structure of Potato Multicystatin-P212121 | | Descriptor: | MULTICYSTATIN | | Authors: | Nissen, M.S, Kumar, G.N, Youn, B, Knowles, D.B, Lam, K.S, Ballinger, W.J, Knowles, N.R, Kang, C. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of Solanum Tuberosum Multicystatin and its Structural Comparison with Other Cystatins.
Plant Cell, 21, 2009
|
|
6AT7
 
 | | Phenylalanine Ammonia-Lyase (PAL) from Sorghum bicolor | | Descriptor: | AMMONIUM ION, Phenylalanine ammonia-lyase | | Authors: | Jun, S.Y, Kang, C. | | Deposit date: | 2017-08-28 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Biochemical and Structural Analysis of Substrate Specificity of a Phenylalanine Ammonia-Lyase.
Plant Physiol., 176, 2018
|
|
3TRQ
 
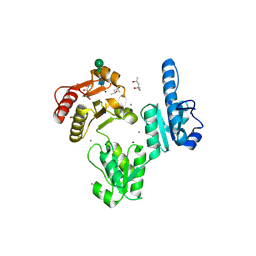 | | Crystal structure of native rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Glycosylation of Skeletal Calsequestrin: IMPLICATIONS FOR ITS FUNCTION.
J.Biol.Chem., 287, 2012
|
|
2J3K
 
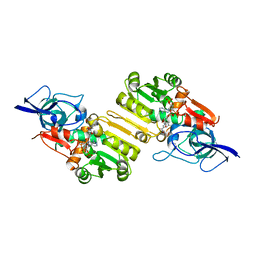 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
3TRP
 
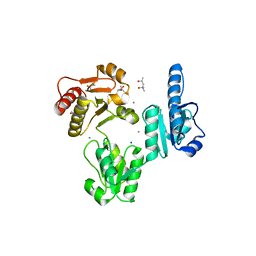 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
2J3J
 
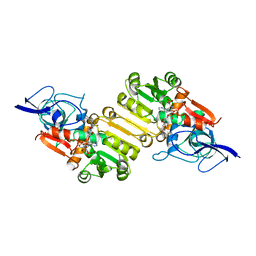 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
1RH1
 
 | | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | | Descriptor: | Colicin B | | Authors: | Hilsenbeck, J.L, Park, H, Chen, G, Youn, B, Postle, K, Kang, C. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution
Mol.Microbiol., 51, 2004
|
|
3V1W
 
 | | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Subramanian, A.K, Nissen, M.N, Lewis, K.M, Sanchez, E.J, Muralidharan, A.K, Kang, C. | | Deposit date: | 2011-12-10 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin
To be Published
|
|
3US3
 
 | | Recombinant rabbit skeletal calsequestrin-MPD complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Nissen, M.S, Munske, G.R, Kang, C. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Glycosylation of skeletal calsequestrin: implications for its function.
J.Biol.Chem., 287, 2012
|
|
2IUK
 
 | | Crystal structure of Soybean Lipoxygenase-D | | Descriptor: | FE (III) ION, SEED LIPOXYGENASE | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
2IUJ
 
 | | Crystal Structure of Soybean Lipoxygenase-B | | Descriptor: | FE (III) ION, LIPOXYGENASE L-5 | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
2ONG
 
 | | Crystal Structure of of limonene synthase with 2-fluorogeranyl diphosphate (FGPP). | | Descriptor: | (1S)-1-[(1R)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ONH
 
 | | Crystal Structure of of limonene synthase with 2-fluorolinalyl diphosphate(FLPP) | | Descriptor: | (1S)-1-[(1S)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4LZI
 
 | | Characterization of Solanum tuberosum Multicystatin and Significance of Core Domains | | Descriptor: | Multicystatin | | Authors: | Nissen, M.S, Kumar, G.N, Green, A.R, Knowles, N.R, Kang, C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Solanum tuberosum Multicystatin and the Significance of Core Domains.
Plant Cell, 25, 2013
|
|
2N7R
 
 | |
2N7Q
 
 | |
4G5E
 
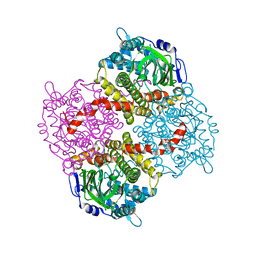 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
4FQU
 
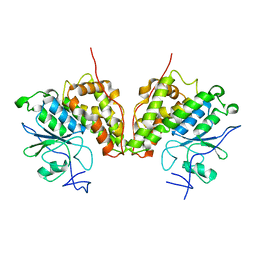 | |
2MXB
 
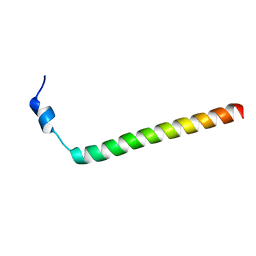 | |
4G0I
 
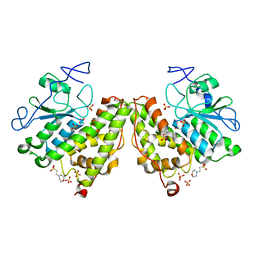 | | Glutathionyl-Hydroquinone Reductase, YqjG of Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, protein yqjG | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G0K
 
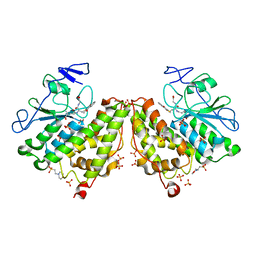 | | Glutathionyl-hydroquinone reductase, YqjG, of E.coli complexed with GS-menadione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-gamma-glutamyl-S-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-L-cysteinylglycine, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G0L
 
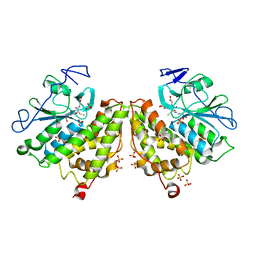 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
