1IZL
 
 | | Crystal Structure of Photosystem II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Kamiya, N, Shen, J.-R. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2EIH
 
 | |
2EHD
 
 | | Crystal Structure Analysis of Oxidoreductase | | Descriptor: | COBALT (II) ION, Oxidoreductase, short-chain dehydrogenase/reductase family | | Authors: | Kamiya, N, Hikima, T, Ebihara, A, Inoue, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of putative oxidoreductase from Thermus thermophilus HB8
to be published
|
|
4TWZ
 
 | | Crystal Structure Analysis of E Coli. RecA Protein | | Descriptor: | MAGNESIUM ION, Protein RecA | | Authors: | Hikima, T, Hiraki, T, Furuse, M, Ikawa, S, Iwasaki, W, Shibata, T, Kamiya, N. | | Deposit date: | 2014-07-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair
Nucleic Acids Res., 43, 2015
|
|
5H2F
 
 | | Crystal structure of the PsbM-deletion mutant of photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Uto, S, Kawakami, K, Umena, Y, Iwai, M, Ikeuchi, M, Shen, J.R, Kamiya, N. | | Deposit date: | 2016-10-15 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual relationships between structural and functional changes in a PsbM-deletion mutant of photosystem II.
Faraday Discuss., 198, 2017
|
|
8ISN
 
 | | HLA-A24 in complex with modified 9mer WT1 peptide | | Descriptor: | Beta-2-microglobulin, CYS-TYR-THR-TRP-ASN-GLN-MET-ASN-LEU, GLYCEROL, ... | | Authors: | Bekker, G.J, Numoto, N, Kawasaki, M, Hayashi, T, Yabuno, S, Kozono, Y, Shimizu, T, Kozono, H, Ito, N, Oda, M, Kamiya, N. | | Deposit date: | 2023-03-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Elucidation of binding mechanism, affinity, and complex structure between mWT1 tumor-associated antigen peptide and HLA-A*24:02.
Protein Sci., 32, 2023
|
|
7CTR
 
 | | Closed form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
7CTS
 
 | | Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein, BICINE, ... | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
1ONL
 
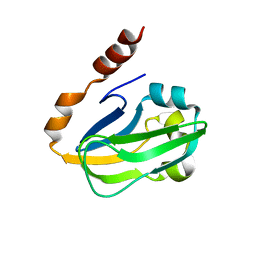 | | Crystal structure of Thermus thermophilus HB8 H-protein of the glycine cleavage system | | Descriptor: | glycine cleavage system H protein | | Authors: | Nakai, T, Ishijima, J, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-28 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Thermus thermophilus HB8 H-protein of the glycine-cleavage system, resolved by a six-dimensional molecular-replacement method.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7VEB
 
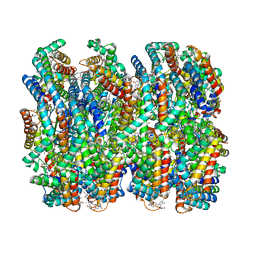 | | Phycocyanin rod structure of cyanobacterial phycobilisome | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Kawakami, K, Hamaguchi, T, Hirose, Y, Kosumi, D, Miyata, M, Kamiya, N, Yonekura, K. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Nat Commun, 13, 2022
|
|
7VEA
 
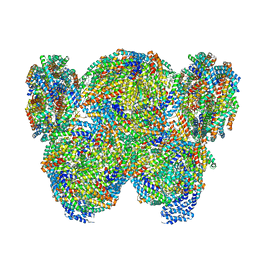 | | Pentacylindrical allophycocyanin core from Thermosynechococcus vulcanus | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Kawakami, K, Hamaguchi, T, Hirose, Y, Kosumi, D, Miyata, M, Kamiya, N, Yonekura, K. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Nat Commun, 13, 2022
|
|
8Z2G
 
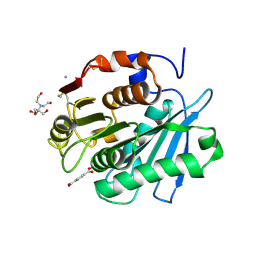 | | MHET bound form of PET-degrading cutinase mutant Cut190*SS_S176A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, AMMONIUM ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2H
 
 | | Substrate analog a010 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[2-hydroxyethyloxy(oxidanyl)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2I
 
 | | Substrate analog a011 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 2-hydroxyethyloxy-(4-methoxycarbonylphenyl)phosphinic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2K
 
 | | Substrate analog a013 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[oxidanyl(2-phenylmethoxyethoxy)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2J
 
 | | Substrate analog a012 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | (4-methoxycarbonylphenyl)-(2-phenylmethoxyethoxy)phosphinic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
1UMD
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UMB
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UMC
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methylpentanoate | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, 4-METHYL VALERIC ACID, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UM9
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in apo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, SULFATE ION | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
8IYT
 
 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Racemization of the substrate and product by serine palmitoyltransferase from Sphingobacterium multivorum yields two enantiomers of the product from d-serine.
J.Biol.Chem., 300, 2024
|
|
8IYP
 
 | | Crystal structure of serine palmitoyltransferase soaked in 190 mM D-serine solution | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Racemization of the substrate and product by serine palmitoyltransferase from Sphingobacterium multivorum yields two enantiomers of the product from d-serine.
J.Biol.Chem., 300, 2024
|
|
1DZE
 
 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
5ZRS
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | Descriptor: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | Authors: | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
