7EWE
 
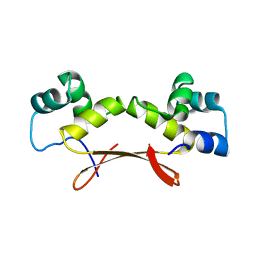 | | Mycobacterium tuberculosis HigA2 (Form III) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7EWD
 
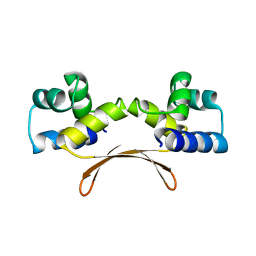 | | Mycobacterium tuberculosis HigA2 (Form II) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
5ZHF
 
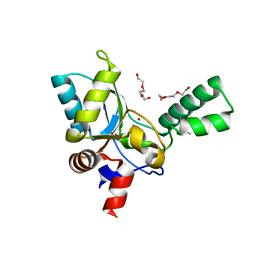 | | Structure of VanYB unbound | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-03-13 | | Release date: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
5ZHW
 
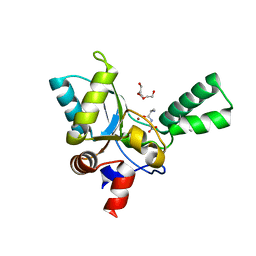 | | VanYB in complex with D-Alanine-D-Alanine | | Descriptor: | COPPER (II) ION, D-ALANINE, D-alanyl-D-alanine carboxypeptidase, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-03-13 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
4LMP
 
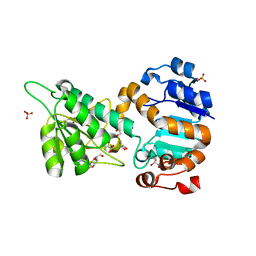 | | Mycobacterium tuberculosis L-alanine dehydrogenase x-ray structure in complex with N6-methyl adenosine | | Descriptor: | Alanine dehydrogenase, GLYCEROL, N-methyladenosine, ... | | Authors: | Kim, H.-B, Hung, L.-W, Goulding, C.W, Terwilliger, T.C, Kim, C.-Y, Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-07-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug target analysis by dye-ligand affinity chromatography
To be Published
|
|
6A6A
 
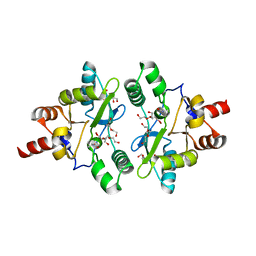 | | VanYB in complex with D-Alanine | | Descriptor: | ACETATE ION, D-ALANINE, D-alanyl-D-alanine carboxypeptidase, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
1A7K
 
 | | GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN A MONOCLINIC CRYSTAL FORM | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Hol, W.G.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Leishmania mexicana glycosomal glyceraldehyde-3-phosphate dehydrogenase in a new crystal form confirms the putative physiological active site structure.
J.Mol.Biol., 278, 1998
|
|
3LP6
 
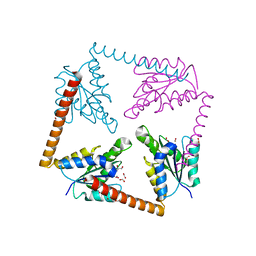 | | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A resolution | | Descriptor: | FORMIC ACID, GLYCEROL, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kim, H, Yu, M, Hung, L.-W, Terwilliger, T.C, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A
Resolution
To be Published
|
|
4HC6
 
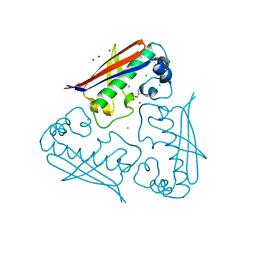 | | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, H.-B, Han, G.-W, Hung, L.-W, Terwilliger, C.T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution
To be Published
|
|
7BWK
 
 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA-VpdB(461-590) derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Oh, B.H. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for effector protein recognition by the Dot/Icm Type IVB coupling protein complex.
Nat Commun, 11, 2020
|
|
8J1O
 
 | | Crystal structure of HaloTag complexed with BTTA | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, N-[[1-[2-[2-(2-hexoxyethoxy)ethoxy]ethyl]-1,2,3-triazol-4-yl]methyl]-1-(1H-1,2,3-triazol-4-yl)-N-(2H-1,2,3-triazol-4-ylmethyl)methanamine, ... | | Authors: | Kim, H, Rhee, H, Lee, C. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of HaloTag complexed with BTTA
To Be Published
|
|
6V88
 
 | |
6MW8
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
6MW5
 
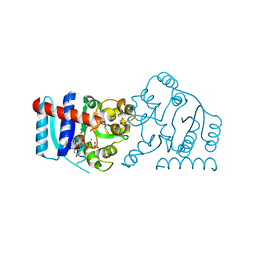 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Platinum | | Descriptor: | 1,2-ETHANEDIOL, PLATINUM (II) ION, UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
3PJF
 
 | |
3PJE
 
 | |
3PJD
 
 | |
6IOZ
 
 | | Structural insights of idursulfase beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, H, Kim, D, Hong, J, Lee, K, Seo, J, Oh, B.H. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights of idursulfase beta
To Be Published
|
|
1A5C
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE FROM PLASMODIUM FALCIPARUM | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Kim, H, Certa, U, Dobeli, H, Jakob, P, Hol, W.G.J. | | Deposit date: | 1998-02-13 | | Release date: | 1998-06-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum.
Biochemistry, 37, 1998
|
|
3AXX
 
 | |
6KQB
 
 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
6KQ9
 
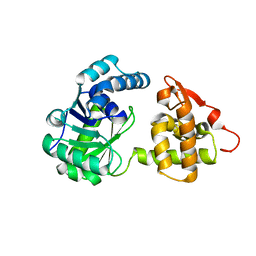 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
1MXJ
 
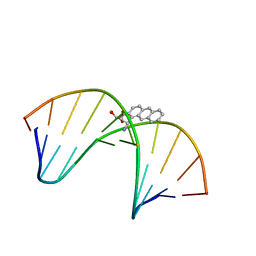 | | NMR solution structure of benz[a]anthracene-dG in ras codon 12,2; GGCAGXTGGTG | | Descriptor: | 1S,2R,3S,4R-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*AP*CP*CP*AP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*G)-3' | | Authors: | Kim, H.-Y.H, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minor Groove Orientation for the (1S,2R,3S,4R)-N2-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxy-benz[a]anthracenyl)]-2'-deoxyguanosyl Adduct in the N-ras Codon 12 sequence
Biochemistry, 42, 2003
|
|
8GHL
 
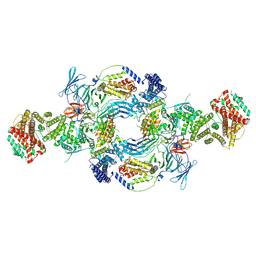 | | the Hir complex core | | Descriptor: | Histone transcription regulator 3, Protein HIR1, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHN
 
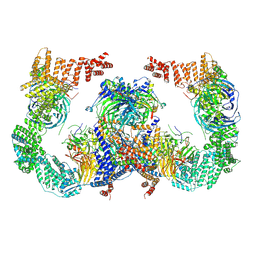 | |
