5W0G
 
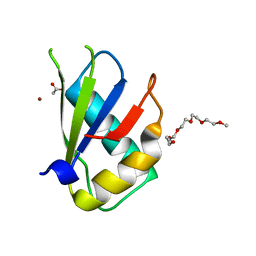 | | Structure of U2AF65 (U2AF2) RRM1 at 1.07 resolution | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, Splicing factor U2AF 65 kDa subunit, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2017-05-30 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs.
Biochemistry, 56, 2017
|
|
5W0H
 
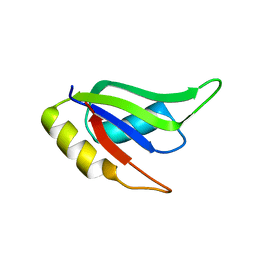 | |
6XLX
 
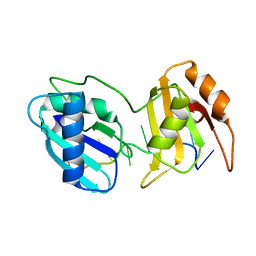 | |
6XLV
 
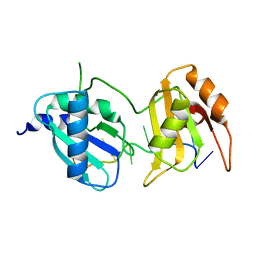 | |
3BS9
 
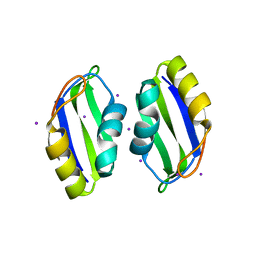 | | X-ray structure of human TIA-1 RRM2 | | Descriptor: | IODIDE ION, Nucleolysin TIA-1 isoform p40 | | Authors: | Kumar, A.O, Kielkopf, C.L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution.
Biochem.Biophys.Res.Commun., 367, 2008
|
|
5EV1
 
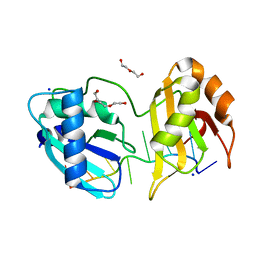 | | Structure I of Intact U2AF65 Recognizing a 3' Splice Site Signal | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*UP*UP*U)-D(P*UP*UP*(BRU)P*U)-R(P*UP*U)-3'), SODIUM ION, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV2
 
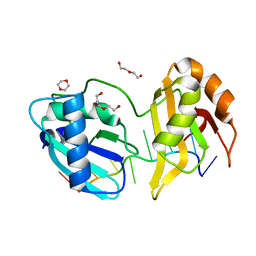 | | Structure II of Intact U2AF65 Recognizing the 3' Splice Site Signal | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, DNA (5'-R(P*UP*U)-D(P*UP*U)-R(P*U)-D(P*UP*(BRU)P*U)-3'), ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV3
 
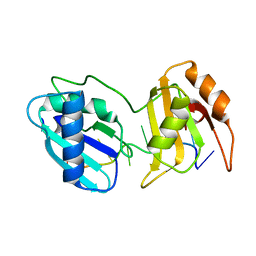 | |
5EV4
 
 | |
6N3D
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif (APO-State) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6NSX
 
 | |
6N3E
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to U2AF ligand motif 4 | | Descriptor: | FORMIC ACID, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6N3F
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to SF3b1 ULM5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Leach, J.R, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
3CGP
 
 | |
3CGS
 
 | |
3CGR
 
 | |
3CGQ
 
 | |
7S3C
 
 | |
7S3B
 
 | |
3VAK
 
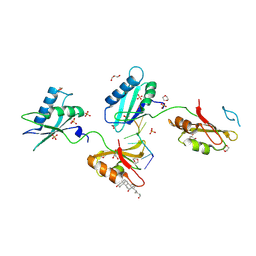 | | Structure of U2AF65 variant with BrU5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAF
 
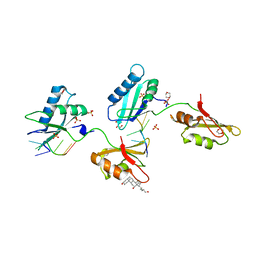 | | Structure of U2AF65 variant with BrU3 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*(BRU)P*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAM
 
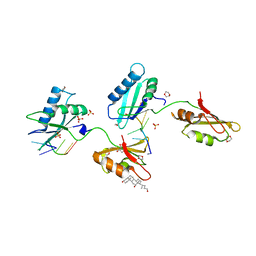 | | Structure of U2AF65 variant with BrU5C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*CP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAJ
 
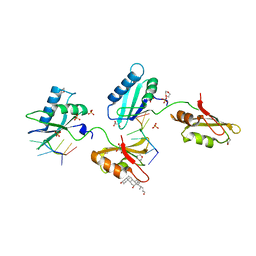 | | Structure of U2AF65 variant with BrU5C6 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*CP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAI
 
 | | Structure of U2AF65 variant with BrU3C5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*UP*CP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAH
 
 | | Structure of U2AF65 variant with BrU3C4 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(P*UP*UP*(BRU)P*CP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
