4ZPI
 
 | | Crystal Structure of HygX from Streptomyces hygroscopicus with iron bound | | Descriptor: | FE (II) ION, Putative oxidase/hydroxylase, SUCCINIC ACID | | Authors: | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3HSC
 
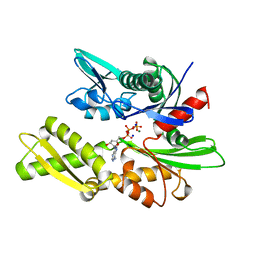 | | THREE-DIMENSIONAL STRUCTURE OF THE ATPASE FRAGMENT OF A 70K HEAT-SHOCK COGNATE PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 7OKD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Deluca-Flaherty, C.R, Mckay, D.B. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein.
Nature, 346, 1990
|
|
6I3Q
 
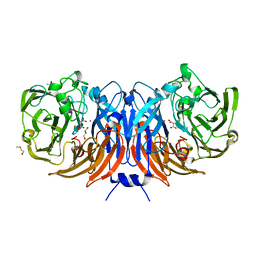 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus complex with acetate ions. | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Polyakov, K.M, Popov, A.N, Tikhkonova, T.V, Popov, V.O, Trofimov, A.A. | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4J6D
 
 | |
5A2A
 
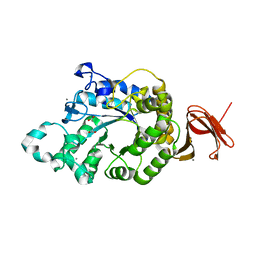 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ACETATE ION, APO FORM OF ANOXYBACILLUS ALPHA-AMYLASES, CALCIUM ION | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-16 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5AM9
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta 10-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, CALCIUM ION, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
5A2B
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5AMB
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta 35-42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMYLOID BETA A4 PROTEIN, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
5AMC
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta fluorogenic fragment 4-10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
5AMA
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta 1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
6HWI
 
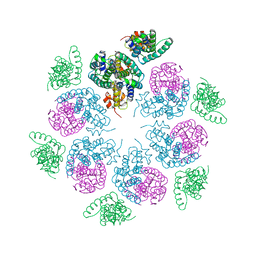 | | Immature M-PMV capsid hexamer structure in intact virus particles | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Qu, K, Glass, B, Dolezal, M, Schur, F.K.M, Rein, A, Rumlova, M, Ruml, T, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HWW
 
 | | Immature MLV capsid hexamer structure in intact virus particles | | Descriptor: | Putative gag polyprotein | | Authors: | Qu, K, Glass, B, Dolezal, M, Schur, F.K.M, Rein, A, Rumlova, M, Ruml, T, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2018-10-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5A2C
 
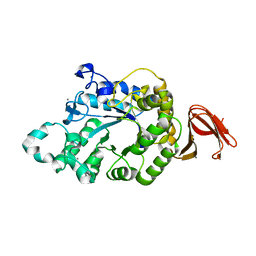 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
6I7D
 
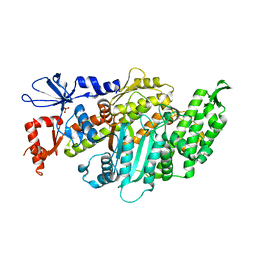 | | Plasmodium falciparum Myosin A, post-rigor and rigor-like states | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Myosin-A | | Authors: | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
3HHP
 
 | |
3JTM
 
 | | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Timofeev, V.I, Shabalin, I.G, Serov, A.E, Polyakov, K.M, Popov, V.O, Tishkov, V.I, Kuranova, I.P, Samigina, V.R. | | Deposit date: | 2009-09-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana
to be published
|
|
5BQF
 
 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
5J8O
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2R)-1-({3-bromo-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)piperidine-2-carboxylic acid, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
5T9T
 
 | | Protocadherin Gamma B2 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin gamma B2-alpha C, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
8R07
 
 | | C-terminal Rel-homology Domain of NFAT1 | | Descriptor: | Nuclear factor of activated T-cells, cytoplasmic 2 | | Authors: | Zak, K.M, Boettcher, J. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ligandability assessment of the C-terminal Rel-homology domain of NFAT1.
Arch Pharm, 357, 2024
|
|
5TME
 
 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
5JMN
 
 | | Fusidic acid bound AcrB | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, DARPin, ... | | Authors: | Oswald, C, Tam, H.K, Pos, K.M. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB.
Nat Commun, 7, 2016
|
|
5JFQ
 
 | | Geranylgeranyl Pyrophosphate Synthetase from archaeon Geoglobus acetivorans | | Descriptor: | Geranylgeranyl Pyrophosphate Synthetase | | Authors: | Petrova, T, Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of geranylgeranyl pyrophosphate synthase GACE1337 from the hyperthermophilic archaeon Geoglobus acetivorans.
Extremophiles, 22, 2018
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
8QPT
 
 | | Crystal structure of pyrophosphatase from Ogataea parapolymorpha | | Descriptor: | GLYCEROL, MAGNESIUM ION, inorganic diphosphatase | | Authors: | Matyuta, I.O, Rodina, E.V, Vorobyeva, N.N, Kurilova, S.A, Bezpalaya, E.Y, Boyko, K.M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast mitochondrial type pyrophosphatase provides a model to study pathological mutations in its human ortholog.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
