3COC
 
 | |
3COD
 
 | | Crystal Structure of T90A/D115A mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Joh, N.H, Min, A, Faham, S, Bowie, J.U. | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins.
Nature, 453, 2008
|
|
4P6L
 
 | |
4P6J
 
 | |
4P6K
 
 | |
3HAR
 
 | |
3HAP
 
 | | Crystal structure of bacteriorhodopsin mutant L111A crystallized from bicelles | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Joh, N.H, Yang, D, Bowie, J.U. | | Deposit date: | 2009-05-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Similar energetic contributions of packing in the core of membrane and water-soluble proteins.
J.Am.Chem.Soc., 131, 2009
|
|
3HAS
 
 | | Crystal structure of bacteriorhodopsin mutant L152A crystallized from bicelles | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Joh, N.H, Yang, D, Bowie, J.U. | | Deposit date: | 2009-05-02 | | Release date: | 2009-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Similar energetic contributions of packing in the core of membrane and water-soluble proteins.
J.Am.Chem.Soc., 131, 2009
|
|
3HAQ
 
 | | Crystal structure of bacteriorhodopsin mutant I148A crystallized from bicelles | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Joh, N.H, Yang, D, Bowie, J.U. | | Deposit date: | 2009-05-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Similar energetic contributions of packing in the core of membrane and water-soluble proteins.
J.Am.Chem.Soc., 131, 2009
|
|
3HAN
 
 | |
3HAO
 
 | |
7M8J
 
 | | SARS-CoV-2 S-NTD + Fab CM25 | | Descriptor: | CM25 Fab - Heavy Chain, CM25 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes.
Science, 372, 2021
|
|
8V5A
 
 | |
8V5K
 
 | | Structure of the Human Respirovirus 3 Fusion Protein Bound to Camelid Nanobodies 4C03 and 4C06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Camelid Nanobody 4C03, Camelid Nanobody 4C06, ... | | Authors: | Johnson, N.V, Ramamohan, A.R, McLellan, J.S. | | Deposit date: | 2023-11-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for potent neutralization of human respirovirus type 3 by protective single-domain camelid antibodies.
Nat Commun, 15, 2024
|
|
8V62
 
 | |
8VCR
 
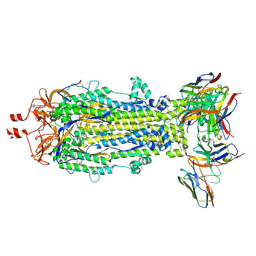 | | SARS-CoV-2 Spike S2 bound to Fab 54043-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 54043-5 Fab Heavy Chain, ... | | Authors: | Johnson, N.V, McLellan, J.S. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and characterization of a pan-betacoronavirus S2-binding antibody.
Structure, 2024
|
|
7T01
 
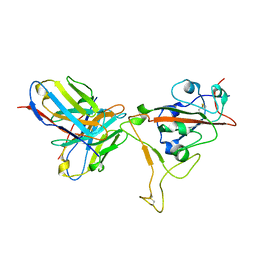 | | SARS-CoV-2 S-RBD + Fab 54042-4 | | Descriptor: | 54042-4 Fab - Heavy Chain, 54042-4 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition.
Cell Rep, 37, 2021
|
|
2MUZ
 
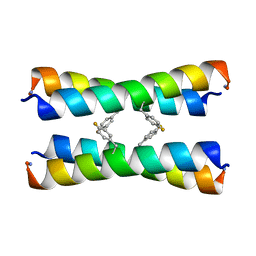 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
5J7D
 
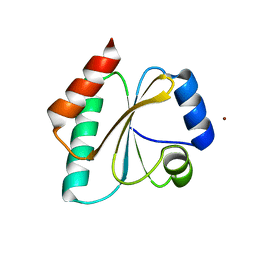 | | Computationally Designed Thioredoxin dF106 | | Descriptor: | COPPER (II) ION, Designed Thioredoxin dF106 | | Authors: | Horowitz, S, Johansen, N, Olsen, J.G, Winther, J.R. | | Deposit date: | 2016-04-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational Redesign of Thioredoxin Is Hypersensitive toward Minor Conformational Changes in the Backbone Template.
J.Mol.Biol., 428, 2016
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
1RCM
 
 | |
1EET
 
 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
2Y8C
 
 | | Plasmodium falciparum dUTPase in complex with a trityl ligand | | Descriptor: | (2S)-2-[(2,4-dioxopyrimidin-1-yl)methyl]-N-(2-hydroxyethyl)-4-trityloxy-butanamide, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, SULFATE ION | | Authors: | Baragana, B, McCarthy, O, Sanchez, P, Bosch, C, Kaiser, M, Brun, R, Whittingham, J.L, Roberts, S, Zhou, X.-X, Wilson, K.S, Johansson, N.G, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2011-02-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beta-Branched Acyclic Nucleoside Analogues as Inhibitors of Plasmodium Falciparum Dutpase
Bioorg.Med.Chem., 19, 2011
|
|
5AR1
 
 | | Crystal structure of Cdc11 from Saccharomyces cerevisiae | | Descriptor: | CELL DIVISION CONTROL PROTEIN 11 | | Authors: | Brausemann, A, Gerhardt, S, Schott, A.K, Einsle, O, Grosse-Berkenbusch, A, Johnsson, N, Gronemeyer, T. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Cdc11, a Septin Subunit from Saccharomyces Cerevisiae.
J.Struct.Biol., 193, 2016
|
|
3T60
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-(tritylamino)uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
