4OBW
 
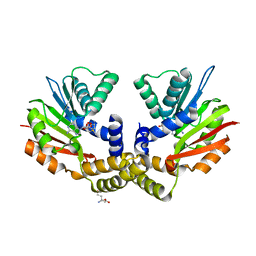 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OI3
 
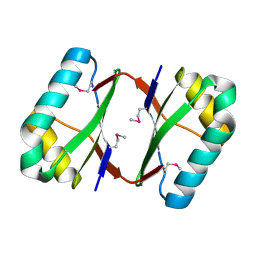 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
5GOQ
 
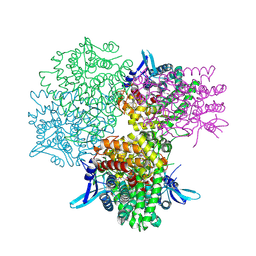 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with glucose | | Descriptor: | Alkaline Invertase, alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5GOO
 
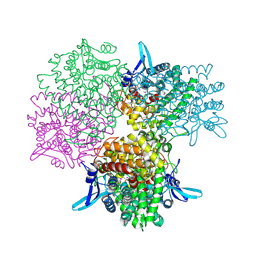 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with fructose | | Descriptor: | Alkaline Invertase, GLYCEROL, beta-D-fructofuranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase.
J. Biol. Chem., 291, 2016
|
|
4PQG
 
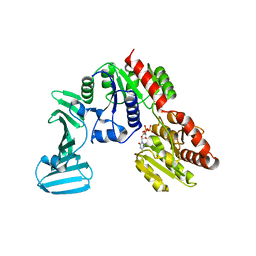 | | Crystal structure of the pneumococcal O-GlcNAc transferase GtfA in complex with UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, URIDINE-5'-DIPHOSPHATE | | Authors: | Shi, W.W, Jiang, Y.L, Zhu, F, Yang, Y.H, Wu, H, Ren, Y.M, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Novel O-Linked N-Acetyl-d-glucosamine (O-GlcNAc) Transferase, GtfA, Reveals Insights into the Glycosylation of Pneumococcal Serine-rich Repeat Adhesins.
J.Biol.Chem., 289, 2014
|
|
5GOR
 
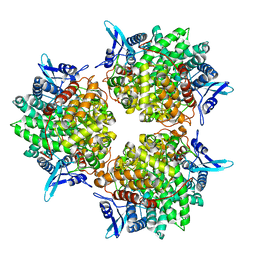 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 | | Descriptor: | Alkaline Invertase, GLYCEROL, SULFATE ION | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.673 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
4Q2W
 
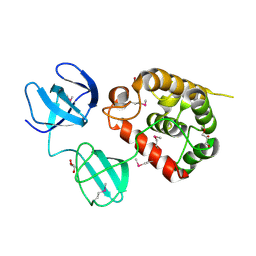 | | Crystal Structure of pneumococcal peptidoglycan hydrolase LytB | | Descriptor: | GLYCEROL, Putative endo-beta-N-acetylglucosaminidase | | Authors: | Bai, X.H, Chen, H.J, Jiang, Y.L, Wen, Z, Cheng, W, Li, Q, Zhang, J.R, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis.
J.Biol.Chem., 289, 2014
|
|
5GOP
 
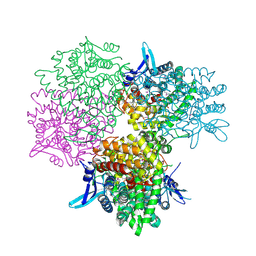 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alkaline Invertase, beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
4QLA
 
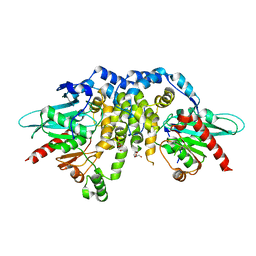 | | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori | | Descriptor: | Juvenile hormone epoxide hydrolase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, K, Jia, N, Hu, C, Jiang, Y.L, Yang, J.P, Chen, Y, Li, S, Zhou, C.Z. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori.
Proteins, 82, 2014
|
|
4R0M
 
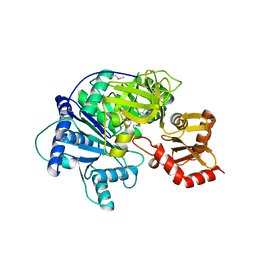 | | Structure of McyG A-PCP complexed with phenylalanyl-adenylate | | Descriptor: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], McyG protein | | Authors: | Tan, X.F, Dai, Y.N, Zhou, K, Jiang, Y.L, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the adenylation-peptidyl carrier protein didomain of the Microcystis aeruginosa microcystin synthetase McyG.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Z8I
 
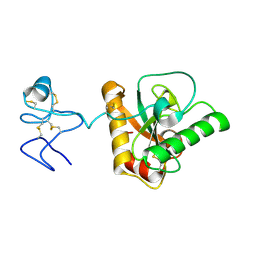 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
4ZXM
 
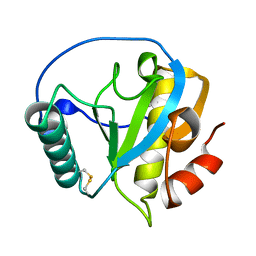 | | Crystal structure of PGRP domain from Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | PGRP domain of peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Yu, H.M, Luo, M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
6J44
 
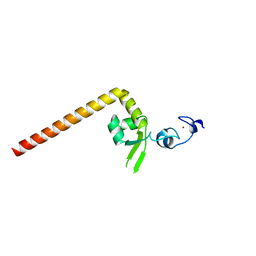 | | Crystal structure of the redefined DNA-binding domain of human XPA | | Descriptor: | DNA repair protein complementing XP-A cells, ZINC ION | | Authors: | Lian, F.M, Yang, X, Yang, W, Jiang, Y.L, Qian, C. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural characterization of the redefined DNA-binding domain of human XPA.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6J3Q
 
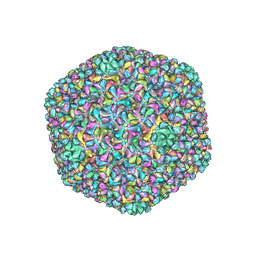 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | Descriptor: | cement protein, major capsid protein | | Authors: | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
6JBJ
 
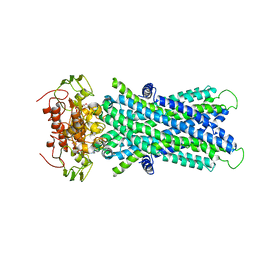 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | Authors: | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
6JYX
 
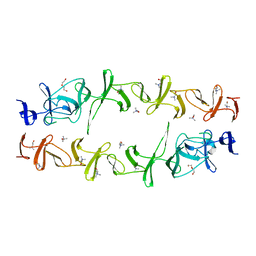 | | Structure of CbpJ from Streptococcus Pneumoniae TIGR4 | | Descriptor: | CHOLINE ION, Choline binding protein J, DI(HYDROXYETHYL)ETHER | | Authors: | Xu, Q, Zhang, J.W, Li, Q, Jiang, Y.L. | | Deposit date: | 2019-04-29 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the choline-binding protein CbpJ from Streptococcus pneumoniae.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6JY5
 
 | | Structure of CsoS4B from Halothiobacillus neapolitanus | | Descriptor: | Unidentified carboxysome polypeptide | | Authors: | Zhao, Y.Y, Jiang, Y.L, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pentameric shell protein CsoS4B of Halothiobacillus neapolitanus alpha-carboxysome.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6KKM
 
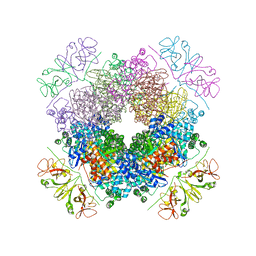 | | Crystal structure of RbcL-Raf1 complex from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6KKN
 
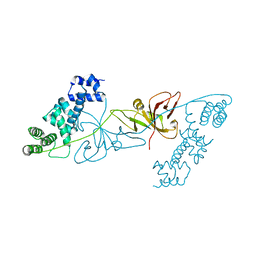 | | Crystal structure of RuBisCO accumulation factor Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6JBH
 
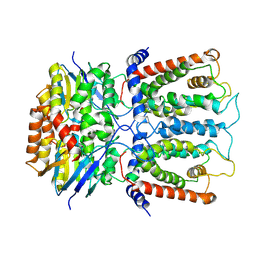 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
6LAE
 
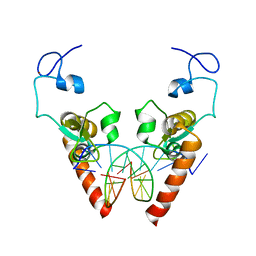 | | Crystal structure of the DNA-binding domain of human XPA in complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*CP*TP*CP*GP*CP*CP*T)-3'), DNA (5'-D(P*TP*GP*GP*CP*GP*AP*GP*AP*TP*GP*C)-3'), DNA repair protein complementing XP-A cells, ... | | Authors: | Lian, F.M, Yang, X, Jiang, Y.L, Yang, F, Li, C, Yang, W, Qian, C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New structural insights into the recognition of undamaged splayed-arm DNA with a single pair of non-complementary nucleotides by human nucleotide excision repair protein XPA.
Int.J.Biol.Macromol., 148, 2020
|
|
8HDS
 
 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDW
 
 | | Cyanophage Pam3 Sheath-tube | | Descriptor: | Pam3 sheath protein, pam3 tube | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
 | |
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
