7W53
 
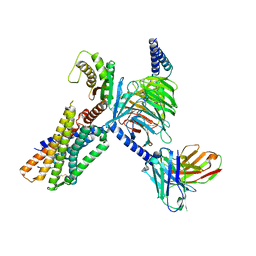 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 1-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
4Z8I
 
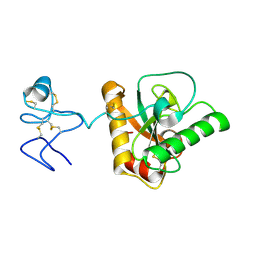 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
7YFW
 
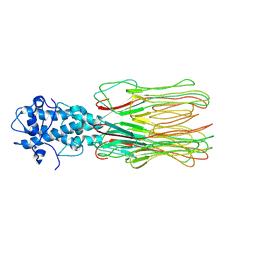 | | Cyanophage Pam3 fiber | | Descriptor: | Pam3 fiber proreins | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6A1K
 
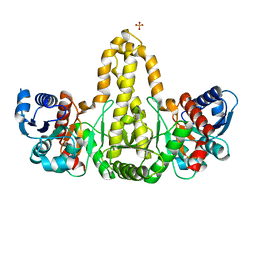 | | Phosphate acyltransferase PlsX from B.subtilis | | Descriptor: | Phosphate acyltransferase, SULFATE ION | | Authors: | Guo, Z, Jiang, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of an amphipathic peptide sensor of the Bacillus subtilisfluid membrane microdomains.
Commun Biol, 2, 2019
|
|
5WPQ
 
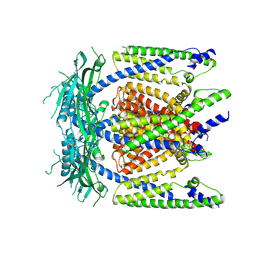 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed I conformation at 3.64 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
5WPV
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs at 3.59 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
5XNJ
 
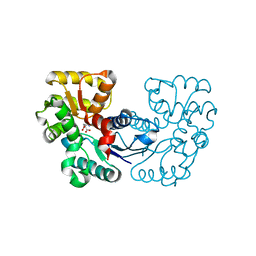 | |
5XNI
 
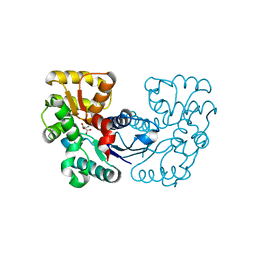 | |
5XNK
 
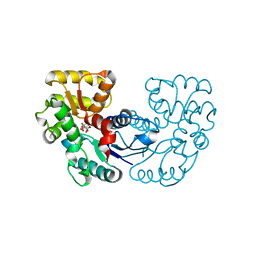 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with DL-methyl-aspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, 3-METHYL-BETA-D-ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function Analyses of a Cyanobacterial Aspartate Racemase Reveal Its Catalytic Mechanism and Substrate Specificity
To Be Published
|
|
