5XBO
 
 | | Lanthanoid tagging via an unnatural amino acid for protein structure characterization | | Descriptor: | Polyubiquitin-B, TERBIUM(III) ION, UV excision repair protein RAD23 homolog A | | Authors: | Jiang, W, Gu, X, Dong, X, Tang, C. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Lanthanoid tagging via an unnatural amino acid for protein structure characterization
J. Biomol. NMR, 67, 2017
|
|
8I79
 
 | | Cryo-EM structure of KCTD7 in complex with Cullin3 | | Descriptor: | BTB/POZ domain-containing protein KCTD7, Cullin-3 | | Authors: | Jiang, W, Wang, W, Zheng, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
7XJL
 
 | | Cryo-EM structure of the spexin-bound GALR2-miniGq complex | | Descriptor: | Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XJK
 
 | |
7XJJ
 
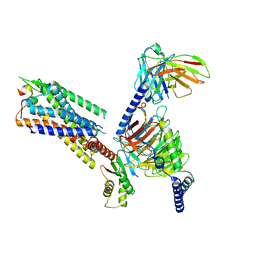 | | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | | Descriptor: | G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Galanin, Galanin receptor type 1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
2XYZ
 
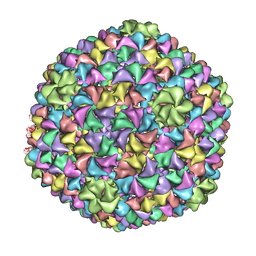 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XYY
 
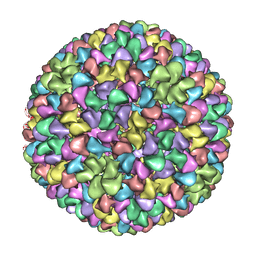 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6R9D
 
 | |
6Y4F
 
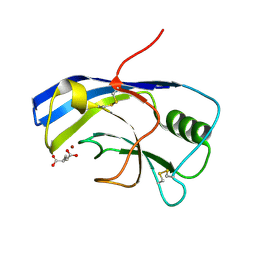 | | X-ray structure of the Zn-dependent receptor-binding domain of Proteus mirabilis MR/P fimbrial adhesin MrpH | | Descriptor: | Fimbrial adhesin, GLUTAMIC ACID, ZINC ION | | Authors: | Knight, S.D, Ubhayasekera, W, Jiang, W. | | Deposit date: | 2020-02-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation.
Plos Pathog., 16, 2020
|
|
6Y4E
 
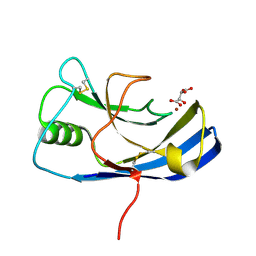 | | X-ray structure of the Zn-dependent receptor-binding domain of Proteus mirabilis MR/P fimbrial adhesin MrpH | | Descriptor: | Fimbrial adhesin, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Knight, S.D, Ubhayasekera, W, Jiang, W. | | Deposit date: | 2020-02-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | MrpH, a new class of metal-binding adhesin, requires zinc to mediate biofilm formation.
Plos Pathog., 16, 2020
|
|
9BRZ
 
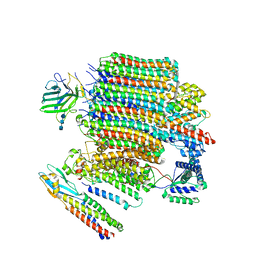 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRU
 
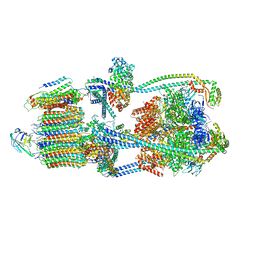 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRT
 
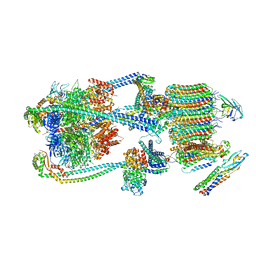 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRR
 
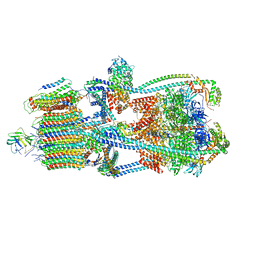 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRS
 
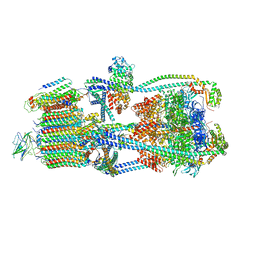 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRY
 
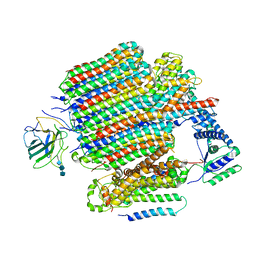 | | V0-only V-ATPase in synaptophysin gene knock-out mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
6WKK
 
 | | Phage G gp27 major capsid proteins and gp26 decoration proteins | | Descriptor: | Gp26 capsid decoration protein, Gp27 major capsid protein | | Authors: | Monroe, L, Gonzalez, B, Jiang, W, Kihara, D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Phage G Structure at 6.1 angstrom Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus.
J.Mol.Biol., 432, 2020
|
|
4WUM
 
 | |
9CVF
 
 | |
9CVG
 
 | |
9CVE
 
 | |
7TMC
 
 | |
