4XE8
 
 | | Bacillus thuringiensis ParM with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2014-12-23 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4XE7
 
 | | Bacillus thuringiensis ParM in apo form | | Descriptor: | Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2014-12-23 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4XHN
 
 | | Bacillus thuringiensis ParM with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2015-01-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4XHO
 
 | | Bacillus thuringiensis ParM with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2015-01-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4XHP
 
 | |
4EMK
 
 | | Crystal structure of SpLsm5/6/7 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5, U6 snRNA-associated Sm-like protein LSm6, U6 snRNA-associated Sm-like protein LSm7 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
4EMG
 
 | | Crystal structure of SpLsm3 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
4EMH
 
 | | Crystal structure of SpLsm4 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm4 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
4JAH
 
 | |
4JAB
 
 | |
4KW0
 
 | | Structure of Dickerson-Drew Dodecamer with 2'-MeSe-ara-G modification | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*(1TW)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Jiang, S, Gan, J, Sheng, J, Sun, H, Huang, Z. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Dickerson-Drew Dodecamer with 2'-MeSe-ara-G modification
TO BE PUBLISHED
|
|
8GU0
 
 | | Crystal structure of a fungal halogenase RadH | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Non-heme halogenase radH, ... | | Authors: | Jiang, S.M, Brown, C.J. | | Deposit date: | 2022-09-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Further Characterization of Fungal Halogenase RadH and Its Homologs.
Biomolecules, 13, 2023
|
|
1RSF
 
 | |
4O0I
 
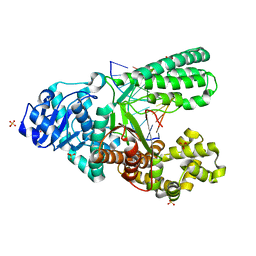 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with 2'-MeSe-arabino-guanosine derivatized DNA | | Descriptor: | 5'-D(*C*(1TW)P*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', DNA polymerase I, GLYCEROL, ... | | Authors: | Jiang, S, Gan, J, Sun, H, Huang, Z. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with 2'-MeSe-arabino-guanosine derivatized DNA
TO BE PUBLISHED
|
|
2NPL
 
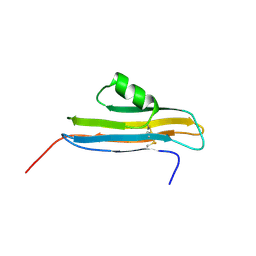 | | NMR Structure of CARD d2 Domain | | Descriptor: | Coxsackievirus and Adenovirus Receptor | | Authors: | Jiang, S, Caffrey, M. | | Deposit date: | 2006-10-27 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coxsackievirus and adenovirus receptor domain 2
Protein Sci., 16, 2007
|
|
8KFX
 
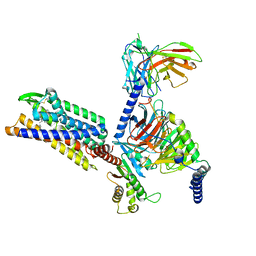 | | Gi bound CCR8 complex with nonpeptide agonist LMD-009 | | Descriptor: | 8-[[3-(2-methoxyphenoxy)phenyl]methyl]-1-(2-phenylethyl)-1,3,8-triazaspiro[4.5]decan-4-one, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFZ
 
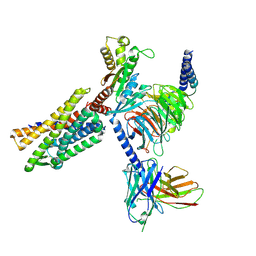 | | Gi bound CCR8 in ligand free state | | Descriptor: | C-C chemokine receptor type 8,LgBiT fusion protein,Recombinant Human Rhinovirus, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFY
 
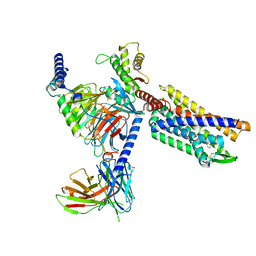 | | Gi bound CCR8 complex with nonpeptide agonist ZK 756326 | | Descriptor: | 2-[2-[4-[(3-phenoxyphenyl)methyl]piperazin-1-yl]ethoxy]ethanol, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
6KZU
 
 | |
3S49
 
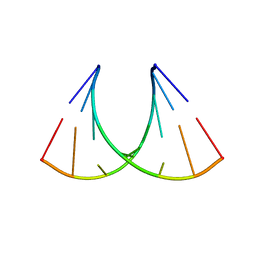 | | RNA crystal structure with 2-Se-uridine modification | | Descriptor: | POTASSIUM ION, RNA (5'-R(*GP*UP*AP*UP*AP*(RUS)P*AP*C)-3') | | Authors: | Sheng, J, Gan, J, Sun, H, Hassan, A.E.H, Jiang, S, Huang, Z. | | Deposit date: | 2011-05-19 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Higher Specificity of RNA Base Pairing with 2-Selenouridine
To be Published
|
|
3OZ9
 
 | |
8HS2
 
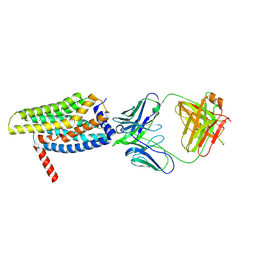 | | Orphan GPR20 in complex with Fab046 | | Descriptor: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS3
 
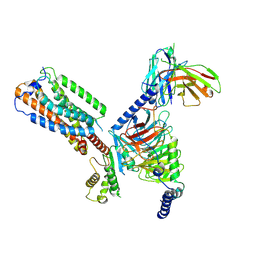 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HSC
 
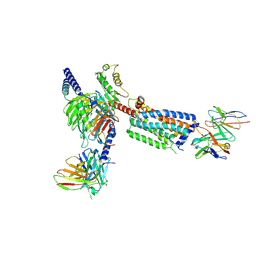 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
1WM7
 
 | | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch, 9 structures | | Descriptor: | Neurotoxin BmP01 | | Authors: | Wu, G, Li, Y, Wei, D, He, F, Jiang, S, Hu, G, Wu, H, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 276, 2000
|
|
