1MP4
 
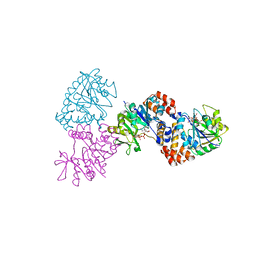 | | W224H VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, W224H Variant of S. Enterica RmlA Bound to UDP-Glucose | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
5VVX
 
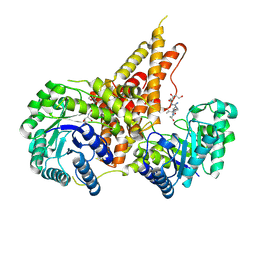 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lamin B1, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
5VVO
 
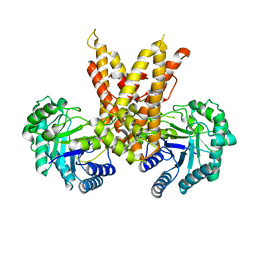 | |
5VVU
 
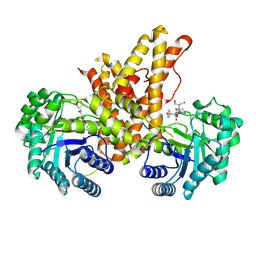 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, TAB1 peptide | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
8D9N
 
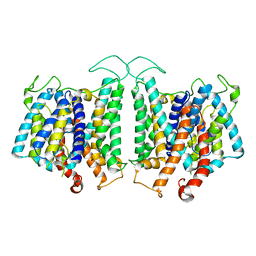 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-06-10 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8E34
 
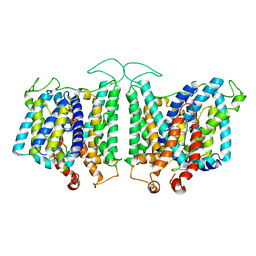 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
7RHQ
 
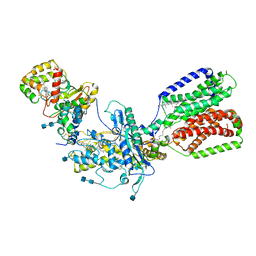 | | Cryo-EM structure of Xenopus Patched-1 in complex with GAS1 and Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, P, Lian, T, Wierbowski, B, Garcia-Linares, S, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
7CJG
 
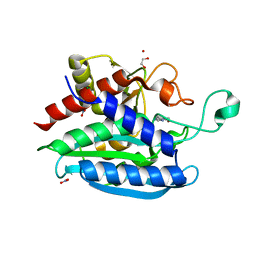 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | 5,6-DIMETHYLBENZIMIDAZOLE, GLYCEROL, Glutamine cyclotransferase-related protein, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
5L77
 
 | | A glycoside hydrolase mutant with an unreacted activity based probe bound | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
6E37
 
 | | O-GlcNAc Transferase in complex with covalent inhibitor | | Descriptor: | (2S,3R,4R,5S,6R)-3-[(2E)-but-2-enoylamino]-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), O-GlcNAc transferase subunit p110, TYR-PRO-GLY-GLY-SER-THR-PRO-VAL-SER-SER-ALA-ASN | | Authors: | Li, H, Jiang, J. | | Deposit date: | 2018-07-13 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Targeted covalent inhibition of O-GlcNAc transferase in cells.
Chem.Commun.(Camb.), 55, 2019
|
|
6WQZ
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
7C72
 
 | | Structure of a mycobacterium tuberculosis puromycin-hydrolyzing peptidase | | Descriptor: | D-MALATE, GLYCEROL, Prolyl oligopeptidase | | Authors: | Ruiz-Carrillo, D, Zhao, Y.H, Feng, Q, Zhou, X, Zhang, Y, Jiang, J, Lukman, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00004458 Å) | | Cite: | Mycobacterium tuberculosis puromycin hydrolase displays a prolyl oligopeptidase fold and an acyl aminopeptidase activity.
Proteins, 89, 2021
|
|
7CJE
 
 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | GLYCEROL, Glutamine cyclotransferase-related protein, MAGNESIUM ION, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.950007 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
3TWH
 
 | | Selenium Derivatized RNA/DNA Hybrid in complex with RNase H Catalytic Domain D132N Mutant | | Descriptor: | DNA (5'-D(*AP*TP*(SDG)P*TP*CP*(SDG))-3'), MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rob, A, Gerlits, O, Jiang, J.S, Gan, J.H, Huang, Z. | | Deposit date: | 2011-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Novel complex MAD phasing and RNase H structural insights using selenium oligonucleotides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SJ1
 
 | | Structure of shaker-W434F | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
7SIP
 
 | | Structure of shaker-IR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
6OSL
 
 | |
3RAU
 
 | | Crystal structure of the HD-PTP Bro1 domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Mu, R.L, Jiang, J.S, Snyder, G, Smith, P, Xiao, T. | | Deposit date: | 2011-03-28 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phe105 Loop of Alix Bro1 Domain Plays a Key Role in HIV-1 Release.
Structure, 19, 2011
|
|
3R9M
 
 | | Crystal structure of the Brox Bro1 domain | | Descriptor: | 1,2-ETHANEDIOL, BRO1 domain-containing protein BROX, FORMIC ACID | | Authors: | Mu, R.L, Jiang, J.S, Snyder, G, Smith, P, Xiao, T. | | Deposit date: | 2011-03-25 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phe105 Loop of Alix Bro1 Domain Plays a Key Role in HIV-1 Release.
Structure, 19, 2011
|
|
4GZ6
 
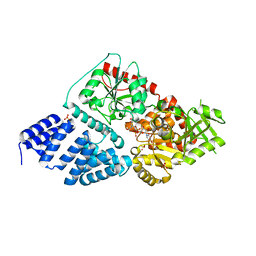 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
7RHR
 
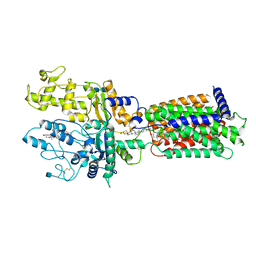 | | Cryo-EM structure of Xenopus Patched-1 in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Huang, P, Lian, T, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
4GZ5
 
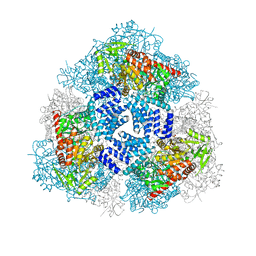 | | Crystal structure of human O-GlcNAc Transferase with UDP-GlcNAc | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
6TYI
 
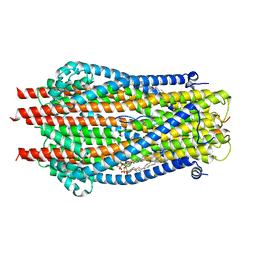 | | ExbB-ExbD complex in MSP1E3D1 nanodisc | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Biopolymer transport protein ExbB, ... | | Authors: | Celia, H, Botos, I, Jiang, J, Buchanan, S.K. | | Deposit date: | 2019-08-09 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry.
Commun Biol, 2, 2019
|
|
4IFP
 
 | | X-ray Crystal Structure of Human NLRP1 CARD Domain | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Jin, T, Curry, J, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Proteins, 81, 2013
|
|
