8TQ8
 
 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ9
 
 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | Descriptor: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQA
 
 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
4DOM
 
 | |
5WCU
 
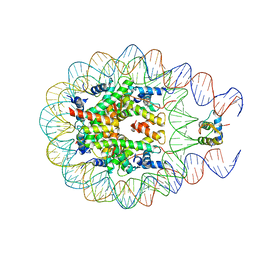 | |
5WER
 
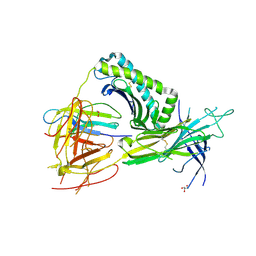 | | Crystal Structure of TAPBPR and H2-Dd complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
6CT0
 
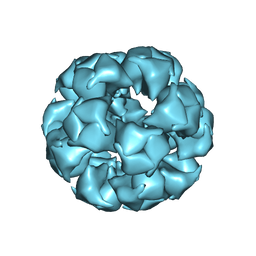 | | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Jiang, J, Baiesc, F.L, Hiromasa, Y, Yu, X, Hui, W.H, Dai, X, Roche, T.E, Zhou, Z.H. | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex.
Biochemistry, 57, 2018
|
|
5WEU
 
 | | Crystal Structure of H2-Dd with disulfide-linked 10mer peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Envelope glycoprotein gp160, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WET
 
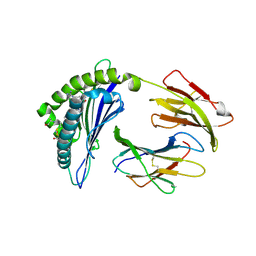 | | Crystal Structure of H2-Dd with disulfide-linked 6mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WES
 
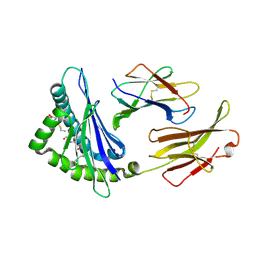 | | Crystal Structure H2-Dd with disulfide-linked 5mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
2GPX
 
 | |
2H05
 
 | |
2HC7
 
 | | 2'-selenium-T A-DNA [G(TSe)GTACAC] | | Descriptor: | 5'-D(*GP*(2ST)P*GP*TP*AP*CP*AP*C)-3' | | Authors: | Jiang, J, Sheng, J, Huang, Z. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of 2'-Selenium modified T A-DNA G(TSe)GTACAC
TO BE PUBLISHED
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3O
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | Descriptor: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
6WR4
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
4OI7
 
 | | RAGE recognizes nucleic acids and promotes inflammatory responses to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*TP*AP*GP*CP*AP*TP*CP*GP*TP*TP*GP*CP*AP*G)-3', 5'-D(*CP*TP*GP*CP*AP*AP*CP*GP*AP*TP*GP*CP*TP*AP*CP*GP*AP*AP*CP*GP*TP*G)-3', ... | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
4OI8
 
 | | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. | | Descriptor: | 5'-D(*CP*CP*AP*TP*GP*AP*CP*TP*GP*TP*AP*GP*GP*AP*AP*AP*CP*TP*CP*TP*AP*GP*A)-3', 5'-D(*CP*TP*CP*TP*AP*GP*AP*GP*TP*TP*TP*CP*CP*TP*AP*CP*AP*GP*TP*CP*AP*TP*G)-3', Advanced glycosylation end product-specific receptor | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
1MP5
 
 | | Y177F VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, Y177F VARIANT OF S. ENTERICA RmlA BOUND TO UDP-GLUCOSE | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
