8DDZ
 
 | | TEM-1 beta-lactamase A237Y | | Descriptor: | Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DE1
 
 | |
8DE2
 
 | | TEM-1 beta-lactamase A237Y mutant covalently bound to avibactam, a room temperature structure | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DE0
 
 | | TEM-1 beta-lactamase covalently bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
7U6Q
 
 | | TEM-1 beta-lactamase | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-03-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
7XRW
 
 | | Solution structure of T. brucei RAP1 | | Descriptor: | Repressor activator protein 1 | | Authors: | Yang, X, Pan, X.H, Ji, Z.Y, Wong, K.B, Zhang, M.J, Zhao, Y.X. | | Deposit date: | 2022-05-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.
Nat Commun, 14, 2023
|
|
6OAA
 
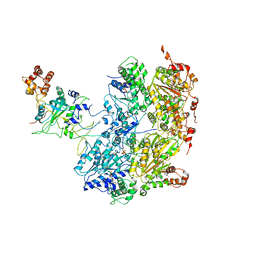 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OA9
 
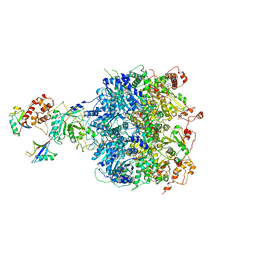 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OAB
 
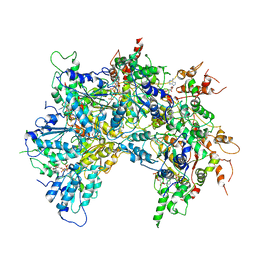 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6J68
 
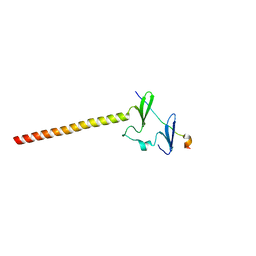 | | Structure of KIBRA and LATS1 Complex | | Descriptor: | Peptide from Serine/threonine-protein kinase LATS1, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJY
 
 | | Crystal Structure of KIBRA and beta-Dystroglycan | | Descriptor: | Peptide from Dystroglycan, Protein KIBRA, SULFATE ION | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJZ
 
 | | Crystal Structure of MAGI2 and Dendrin complex | | Descriptor: | ACETATE ION, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Lin, Z, Zhang, H, Yang, Z, Ji, Z, Zhang, M, Zhu, J. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JJX
 
 | | Crystal Structure of KIBRA and Angiomotin complex | | Descriptor: | Peptide from Angiomotin, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JK0
 
 | | Crystal Structure of YAP1 and Dendrin complex | | Descriptor: | CALCIUM ION, Transcriptional coactivator YAP1,Dendrin | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6J69
 
 | | Structure of KIBRA and Dendrin Complex | | Descriptor: | Peptide from Dendrin, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Kibra Modulates Learning and Memory via Binding to Dendrin.
Cell Rep, 26, 2019
|
|
6JJW
 
 | | Crystal Structure of KIBRA and PTPN14 complex | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6JK1
 
 | | Crystal Structure of YAP1 and Dendrin complex 2 | | Descriptor: | Dendrin,Transcriptional coactivator YAP1 | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
