8I33
 
 | |
1KU0
 
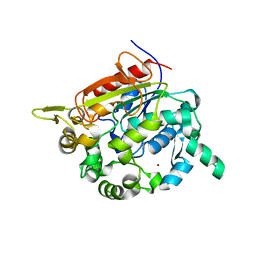 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
6IY0
 
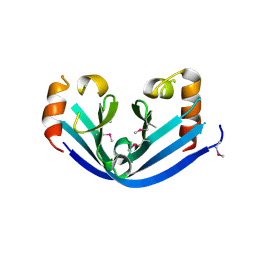 | |
1ZZW
 
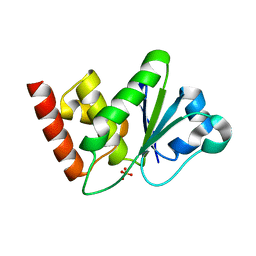 | | Crystal Structure of catalytic domain of Human MAP Kinase Phosphatase 5 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 10, SULFATE ION | | Authors: | Jeong, D.G, Yoon, T.S, Kim, J.H, Shim, M.Y, Jeong, S.K, Son, J.H, Ryu, S.E, Kim, S.J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human MAP Kinase Phosphatase 5: Structural Insight into Constitutively Active Phosphatase.
J.Mol.Biol., 360, 2006
|
|
1YZ4
 
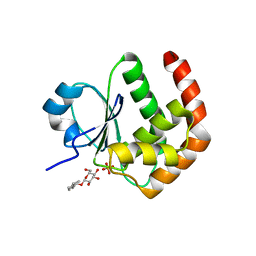 | | Crystal structure of DUSP15 | | Descriptor: | SULFATE ION, dual specificity phosphatase-like 15 isoform a, octyl beta-D-glucopyranoside | | Authors: | Kim, S.J, Ryu, S.E, Jeong, D.G, Yoon, T.S, Kim, J.H, Cho, Y.H, Jeong, S.K, Lee, J.W, Son, J.H. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the catalytic domain of human VHY, a dual-specificity protein phosphatase
Proteins, 61, 2005
|
|
5XZ0
 
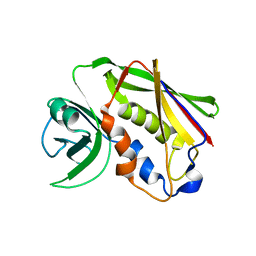 | | Staphylococcal Enterotoxin B (SEB) mutant S19 - N23A, Y90A, R110A and F177A | | Descriptor: | Staphylococcal enterotoxin B | | Authors: | Jeong, W.H, Song, D.H, Hur, G.H, Jeong, S.T. | | Deposit date: | 2017-07-11 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structure of the staphylococcal enterotoxin B vaccine candidate S19 showing eliminated superantigen activity
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5V7I
 
 | | Crystal structure of homo sapiens serine hydroxymethyltransferase 2 (mitochondrial) (SHMT2), in complex with glycine, PLP and folate-competitive pyrazolopyran inhibitor: 6-amino-4-isopropyl-3-methyl-4-(3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl)-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4R)-6-amino-3-methyl-4-(propan-2-yl)-4-[3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl]-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ducker, G.S, Ghergurovich, J.M, Mainolfi, N, Suri, V, Jeong, S, Friedman, A, Manfredi, M, Kim, H, Rabinowitz, J.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2DUV
 
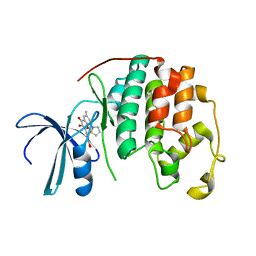 | | Structure of CDK2 with a 3-hydroxychromones | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)-8-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-3-HYDROXY-6-METHYL-4H-CHROMEN-4-ONE, Cell division protein kinase 2 | | Authors: | Kim, K.H, Lee, J, Park, T, Jeong, S, Hong, C. | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Hydroxychromones as cyclin-dependent kinase inhibitors: synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2R64
 
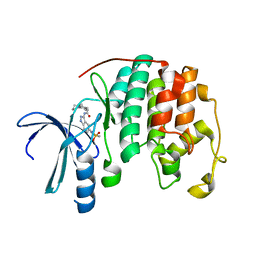 | | Crystal structure of a 3-aminoindazole compound with CDK2 | | Descriptor: | Cell division protein kinase 2, N-[5-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-1H-INDAZOL-3-YL]-2-(4-PIPERIDIN-1-YLPHENYL)ACETAMIDE | | Authors: | Lee, J, Choi, H, Kim, K.H, Jeong, S, Park, J.W, Baek, C.S, Lee, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and biological evaluation of 3,5-diaminoindazoles as cyclin-dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3S2P
 
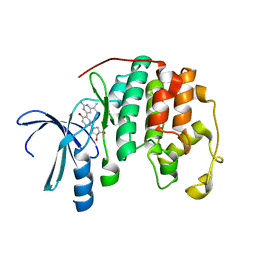 | | Crystal structure of CDK2 with a 2-aminopyrimidine compound | | Descriptor: | (3S,4S)-1-{3-[2-amino-6-(propan-2-yl)pyrimidin-4-yl]-4-hydroxyphenyl}pyrrolidine-3,4-diol, Cyclin-dependent kinase 2 | | Authors: | Kim, K.-H, Lee, J, Jeong, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of 2-aminopyrimidines as CDK1 and CDK2 inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6KUI
 
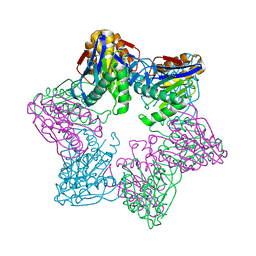 | |
6KWW
 
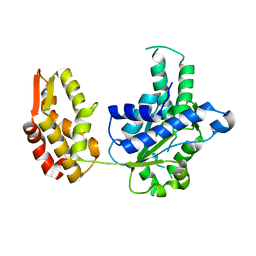 | | HslU from Staphylococcus aureus | | Descriptor: | ATP-dependent protease ATPase subunit HslU | | Authors: | Ha, N.-C, Jeong, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleavage-Dependent Activation of ATP-Dependent Protease HslUV from Staphylococcus aureus .
Mol.Cells, 43, 2020
|
|
6KR1
 
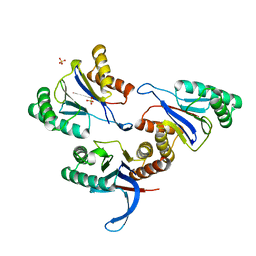 | | ATP dependent protease HslV from Staphylococcus aureus | | Descriptor: | ATP-dependent protease subunit HslV, SULFATE ION | | Authors: | Ha, N.-C, Jeong, S. | | Deposit date: | 2019-08-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cleavage-Dependent Activation of ATP-Dependent Protease HslUV from Staphylococcus aureus .
Mol.Cells, 43, 2020
|
|
5ZNX
 
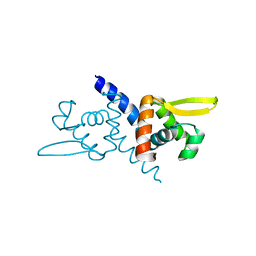 | | Crystal structure of CM14-treated HlyU from Vibrio vulnificus | | Descriptor: | Transcriptional activator | | Authors: | Park, N, Kim, S, Jo, I, Ahn, J, Hong, S, Jeong, S, Baek, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Small-molecule inhibitor of HlyU attenuates virulence of Vibrio species.
Sci Rep, 9, 2019
|
|
7CT8
 
 | | Crystal structure of apo CmoB from Vibrio Vulnificus | | Descriptor: | tRNA U34 carboxymethyltransferase | | Authors: | Kim, J, Jeong, S. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural snapshots of CmoB in various states during wobble uridine modification of tRNA.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7CT9
 
 | | Crystal structure of SAH bound CmoB from Vibrio Vulnificus | | Descriptor: | MALONATE ION, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Kim, J, Jeong, S. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural snapshots of CmoB in various states during wobble uridine modification of tRNA.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7CTA
 
 | | Crystal structure of Cx-SAM bound CmoB from Vibrio vulnificus | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA U34 carboxymethyltransferase | | Authors: | Kim, J, Jeong, S. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural snapshots of CmoB in various states during wobble uridine modification of tRNA.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
