8ARV
 
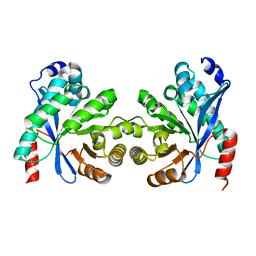 | |
8QNQ
 
 | |
8QNL
 
 | |
2L74
 
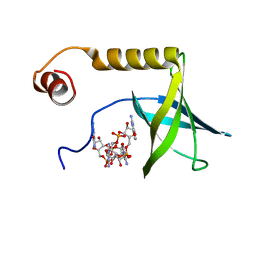 | | Solution structure of the PilZ domain protein PA4608 complex with c-di-GMP identifies charge clustering as molecular readout | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein PA4608 | | Authors: | Habazettl, J, Allan, M, Jenal, U, Grzesiek, S. | | Deposit date: | 2010-12-02 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout
J.Biol.Chem., 286, 2011
|
|
1W25
 
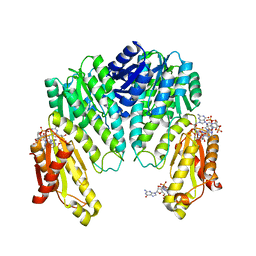 | | Response regulator PleD in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, STALKED-CELL DIFFERENTIATION CONTROLLING PROTEIN, ... | | Authors: | Chan, C, Schirmer, T, Jenal, U. | | Deposit date: | 2004-06-28 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Activity and Allosteric Control of Diguanylate Cyclase
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6SFT
 
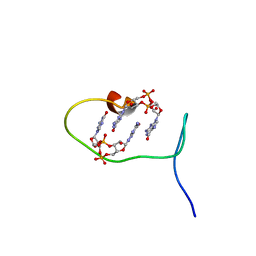 | | Solution structure of protein ARR_CleD in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Two-component receiver protein CleD | | Authors: | Habazettl, J, Hee, C.S, Jenal, U, Schirmer, T, Grzesiek, S. | | Deposit date: | 2019-08-02 | | Release date: | 2020-06-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4AM3
 
 | | Crystal structure of C. crescentus PNPase bound to RNA | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AIM
 
 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AID
 
 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
