1KZY
 
 | | Crystal Structure of the 53bp1 BRCT Region Complexed to Tumor Suppressor P53 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TUMOR SUPPRESSOR P53-BINDING PROTEIN 1, ZINC ION | | Authors: | Joo, W.S, Jeffrey, P.D, Cantor, S.B, Finnin, M.S, Livingston, D.M, Pavletich, N.P. | | Deposit date: | 2002-02-08 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure.
Genes Dev., 16, 2002
|
|
6UFF
 
 | |
5VAF
 
 | |
5VAE
 
 | |
6CFI
 
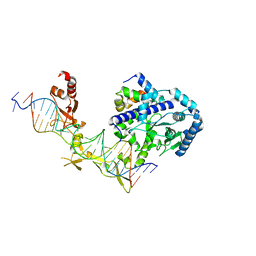 | | Crystal structure of Rad4-Rad23 bound to a 6-4 photoproduct UV lesion | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*(T64)P*TP*GP*GP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), DNA repair protein RAD4, DNA('-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*AP*AP*AP*GP*CP*TP*AP*CP*AP*A)-'), ... | | Authors: | Min, J, Jeffrey, P.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.36241913 Å) | | Cite: | Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex.
Nucleic Acids Res., 47, 2019
|
|
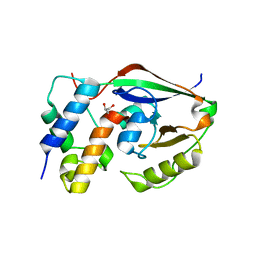
6D0H
 
 | |
6D0I
 
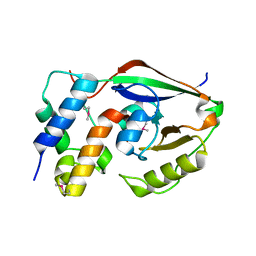 | | ParT: Prs ADP-ribosylating toxin bound to cognate antitoxin ParS. L48M ParT, SeMet-substituted complex. | | Descriptor: | GLYCEROL, ParS: COG5642 (DUF2384) antitoxin fragment, ParT: COG5654 (RES domain) toxin | | Authors: | Piscotta, F.J, Jeffrey, P.D, Link, A.J. | | Deposit date: | 2018-04-10 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ParST is a widespread toxin-antitoxin module that targets nucleotide metabolism.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1TUP
 
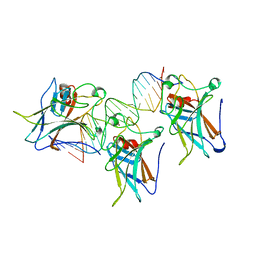 | | TUMOR SUPPRESSOR P53 COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR ), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1995-07-11 | | Release date: | 1995-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
1SHW
 
 | | EphB2 / EphrinA5 Complex Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, Ephrin-A5, ... | | Authors: | Himanen, J.P, Chumley, M.J, Lackmann, M, Li, C, Barton, W.A, Jeffrey, P.D, Vearing, C, Geleick, D, Feldheim, D.A, Boyd, A.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling
Nat.Neurosci., 7, 2004
|
|
1T4W
 
 | | Structural Differences in the DNA Binding Domains of Human p53 and its C. elegans Ortholog Cep-1: Structure of C. elegans Cep-1 | | Descriptor: | C.Elegans p53 tumor suppressor-like transcription factor, ZINC ION | | Authors: | Huyen, Y, Jeffrey, P.D, Derry, W.B, Rothman, J.H, Pavletich, N.P, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Differences in the DNA Binding Domains of Human p53 and Its C. elegans Ortholog Cep-1.
Structure, 12, 2004
|
|
1SHX
 
 | | Ephrin A5 ligand structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-A5 | | Authors: | Himanen, J.P, Barton, W.A, Nikolov, D.B, Jeffrey, P.D. | | Deposit date: | 2004-02-26 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3.
J.Biol.Chem., 280, 2005
|
|
