5L09
 
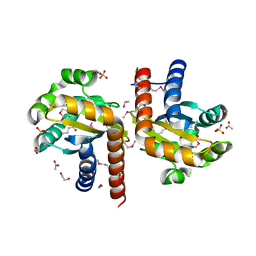 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | 分子名称: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | 著者 | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-07-26 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5L10
 
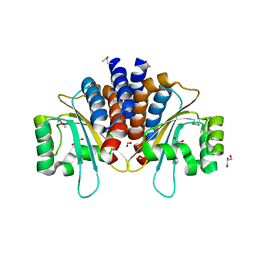 | | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, GLYCEROL, ... | | 著者 | Kim, Y, Chhor, G, Jedrzejczak, R, Winan, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-07-28 | | 公開日 | 2016-10-26 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia.
To Be Published
|
|
6MEL
 
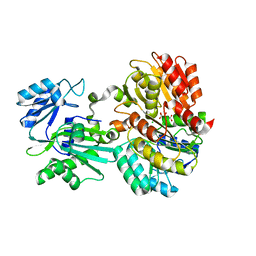 | | Succinyl-CoA synthase from Campylobacter jejuni | | 分子名称: | CHLORIDE ION, CITRIC ACID, Succinate--CoA ligase [ADP-forming] subunit beta, ... | | 著者 | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-09-06 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Succinyl-CoA synthase from Campylobacter jejuni
to be published
|
|
6MGG
 
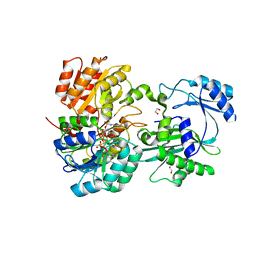 | | Succinyl-CoA synthase from Francisella tularensis, phosphorylated, in complex with CoA | | 分子名称: | 1,2-ETHANEDIOL, COENZYME A, MAGNESIUM ION, ... | | 著者 | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-09-13 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Succinyl-CoA synthase from Francisella tularensis
to be published
|
|
6DIN
 
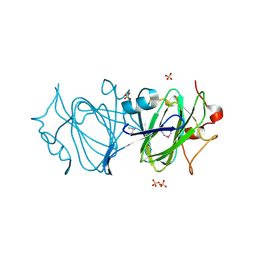 | | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase | | 分子名称: | SULFATE ION, dTDP-4-dehydrorhamnose 3,5-epimerase | | 著者 | Chang, C, Jedrzejczak, R, Chhor, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-23 | | 公開日 | 2018-05-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase
To Be Published
|
|
7KB3
 
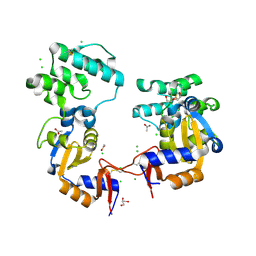 | | The structure of a sensor domain of a histidine kinase (VxrA) from Vibrio cholerae O1 biovar eltor str. N16961, 2nd form | | 分子名称: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-10-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
6C9Z
 
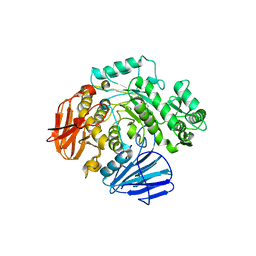 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | 分子名称: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, Glycosyl hydrolase, family 31 | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6C9X
 
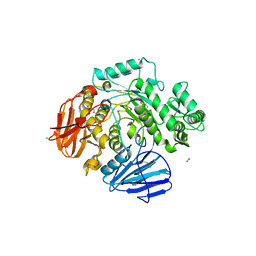 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | 分子名称: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2018-01-29 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.457 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6CA3
 
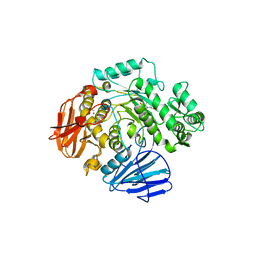 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | 分子名称: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6CA1
 
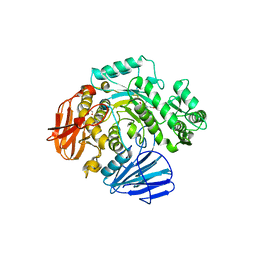 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | 分子名称: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6X1B
 
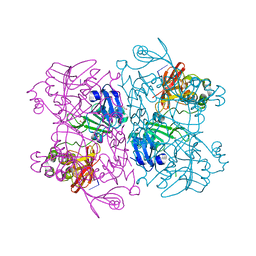 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-18 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WXC
 
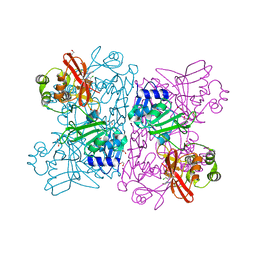 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | 分子名称: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-10 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WLC
 
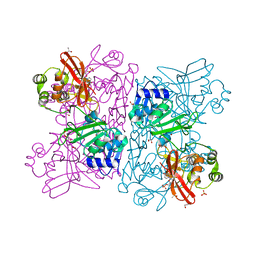 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-19 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5T86
 
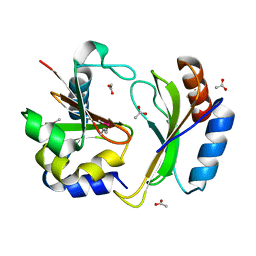 | | Crystal structure of CDI complex from E. coli A0 34/86 | | 分子名称: | ACETATE ION, CdiA toxin, CdiI immunity protein | | 著者 | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-09-06 | | 公開日 | 2017-09-13 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
6WZU
 
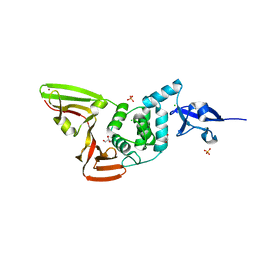 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-14 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6WRH
 
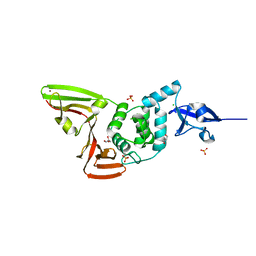 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5KCK
 
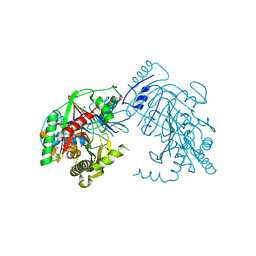 | | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4 | | 分子名称: | Anthranilate synthase component I, GLYCEROL | | 著者 | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-06-06 | | 公開日 | 2016-06-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4
To Be Published
|
|
6XG3
 
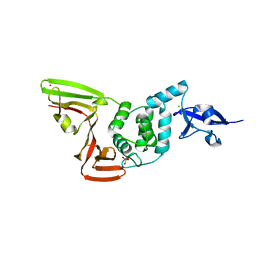 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | 分子名称: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3L1W
 
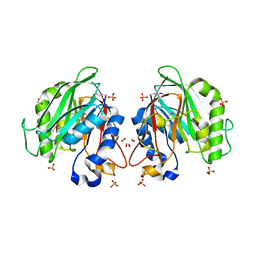 | | The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | 著者 | Tan, K, Rakowski, E, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583
To be Published
|
|
3LOQ
 
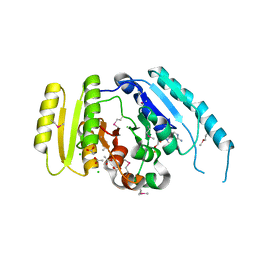 | | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304 | | 分子名称: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | 著者 | Tan, K, Weger, A, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-04 | | 公開日 | 2010-02-16 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | 著者 | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-09-13 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
7KB7
 
 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239-T240 deletion mutant | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | 登録日 | 2020-10-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
7KB9
 
 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, D238-T240 deletion mutant | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Sensor histidine kinase | | 著者 | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-10-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | 分子名称: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | 著者 | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-22 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
