3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1IHD
 
 | |
1JAZ
 
 | |
5M65
 
 | |
1KHQ
 
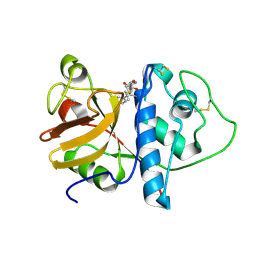 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
5M67
 
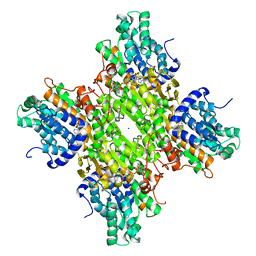 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenine and 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, ACETATE ION, ADENINE, ... | | Authors: | Manszewski, T, Jaskolski, M. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
1JJA
 
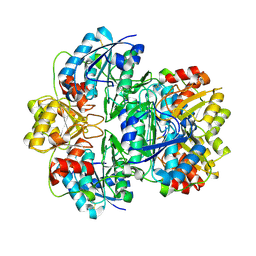 | |
1KHP
 
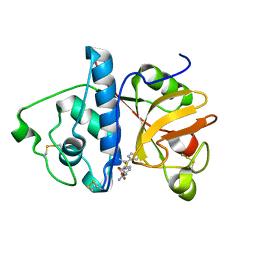 | | Monoclinic form of papain/ZLFG-DAM covalent complex | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
3ONF
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7Z0U
 
 | | Crystal structure of AtWRKY18 DNA-binding domain in complex with W-box DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*GP*GP*CP*G)-3'), WRKY transcription factor 18, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
7Z0R
 
 | | AtWRKY18 DNA-binding domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NITRATE ION, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
6SJJ
 
 | | A new modulated crystal structure of ANS complex of St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, CITRATE ANION, ... | | Authors: | Smietanska, J, Sliwiak, J, Gilski, M, Dauter, Z, Strzalka, R, Wolny, J, Jaskolski, M. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new modulated crystal structure of the ANS complex of the St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4GY9
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3IFA
 
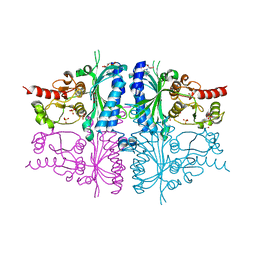 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, GLYCEROL, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3IFC
 
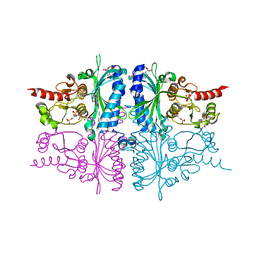 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP and alpha fructose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2NNR
 
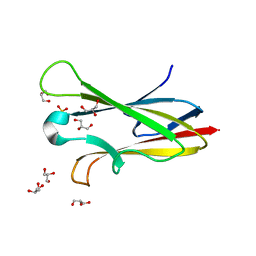 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
5ET5
 
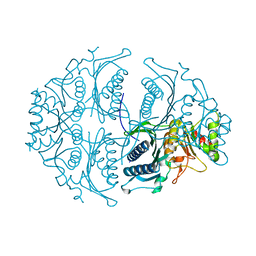 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET8
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | T-to-R switch of muscle FBPase involves extreme changes of secondary and quaternary structure
To Be Published
|
|
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET7
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7QY6
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R5C
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | Authors: | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R1G
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-38 (R207C, D210S, S211V) | | Descriptor: | Beta-aspartyl-peptidase, Isoaspartyl peptidase, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYM
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-18 (R207V, D210P, S211W) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYX
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-24 (R207A, D210S, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
