5NH5
 
 | |
5NHB
 
 | |
5NHA
 
 | |
4V2R
 
 | | Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase | | 分子名称: | PHENYLALANINE AMINOMUTASE (L-BETA-PHENYLALANINE FORMING) | | 著者 | Heberling, M, Masman, M, Bartsch, S, Wybenga, G.G, Dijkstra, B.W, Marrink, S, Janssen, D. | | 登録日 | 2014-10-14 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ironing out their differences: dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase.
ACS Chem. Biol., 10, 2015
|
|
4R9K
 
 | | Structure of thermostable eightfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis | | 分子名称: | (2R)-2-hydroxyhexanamide, GLYCEROL, Limonene-1,2-epoxide hydrolase | | 著者 | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2014-09-05 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
4R9L
 
 | | Structure of a thermostable elevenfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis, containing two stabilizing disulfide bonds | | 分子名称: | (2R)-2-hydroxyhexanamide, Limonene-1,2-epoxide hydrolase | | 著者 | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2014-09-05 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
7AM6
 
 | |
5NH4
 
 | |
8BIT
 
 | | Crystal structure of acyl-CoA synthetase from Metallosphaera sedula in complex with Coenzyme A and acetyl-AMP | | 分子名称: | 4-hydroxybutyrate--CoA ligase 1, COENZYME A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | 著者 | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2022-11-02 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
8BIQ
 
 | | Crystal structure of acyl-COA synthetase from Metallosphaera sedula in complex with acetyl-AMP | | 分子名称: | 4-hydroxybutyrate--CoA ligase 1, ADENOSINE MONOPHOSPHATE, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | 著者 | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2022-11-02 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
6Z1W
 
 | |
6Z1X
 
 | |
7AM3
 
 | |
7AM8
 
 | |
7AM4
 
 | |
7AM5
 
 | |
7AM7
 
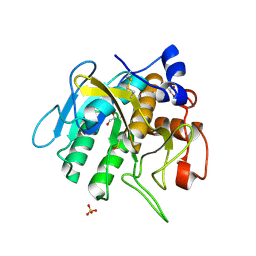 | |
4BAA
 
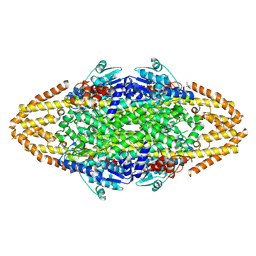 | | Redesign of a Phenylalanine Aminomutase into a beta-Phenylalanine Ammonia Lyase | | 分子名称: | PHENYLALANINE AMMONIA-LYASE | | 著者 | Bartsch, S, Wybenga, G.G, Jansen, M, Heberling, M.M, Wu, B, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2012-09-12 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Redesign of a Phenylalanine Aminomutase Into a Phenylalanine Ammonia Lyase
To be Published
|
|
4BAB
 
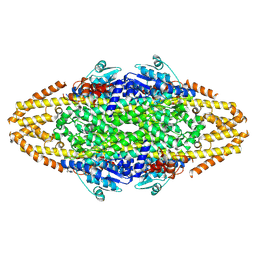 | | Redesign of a Phenylalanine Aminomutase into a beta-Phenylalanine Ammonia Lyase | | 分子名称: | PHENYLALANINE AMINOMUTASE | | 著者 | Bartsch, S, Wybenga, G.G, Jansen, M, Heberling, M.M, Wu, B, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2012-09-12 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Redesign of a Phenylalanine Aminomutase Into a Phenylalanine Ammonia Lyase
To be Published
|
|
7B4J
 
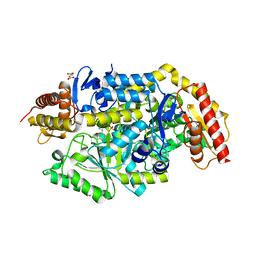 | | Thermostable omega transaminase PjTA-R6 variant W58M/F86L/R417L engineered for asymmetric synthesis of enantiopure bulky amines | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase family protein, SUCCINIC ACID | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2020-12-02 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
7B4I
 
 | | Thermostable omega transaminase PjTA-R6 variant W58G engineered for asymmetric synthesis of enantiopure bulky amines | | 分子名称: | Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE, SUCCINIC ACID | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2020-12-02 | | 公開日 | 2021-09-01 | | 最終更新日 | 2021-09-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
6TP2
 
 | |
6G4C
 
 | |
6G4F
 
 | |
6G4D
 
 | |
