5NHP
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-1-methyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHL
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{R})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
3LAJ
 
 | | The Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound to its DNA Operator and L-arginine. | | Descriptor: | 5'-D(*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*A)-3', 5'-D(*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*A)-3', ARGININE, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2010-01-06 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression.
J.Mol.Biol., 399, 2010
|
|
3LRT
 
 | | Crystal structure of the phosphoribosyl pyrophosphate (PRPP) synthetase from Thermoplasma volcanium in complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Cherney, M.M, Cherney, L.T, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-02-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
5NHH
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHO
 
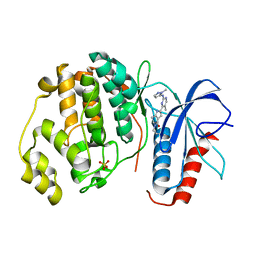 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{S})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHV
 
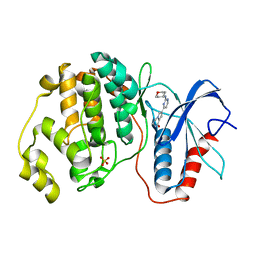 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 7-[2-(oxan-4-ylamino)pyrimidin-4-yl]-3,4-dihydro-2~{H}-pyrrolo[1,2-a]pyrazin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
4TYH
 
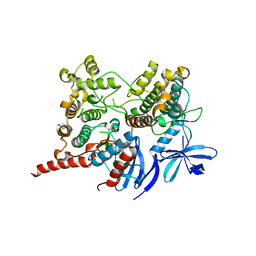 | | Ternary complex of P38 and MK2 with a P38 inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Cumming, J.G, Debreczeni, J.E, Edfeldt, F, Evertsson, F, Harrison, M, Holdgate, G, James, M, Lamont, S, Oldham, K, Sullivan, J.E, Wells, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of substrate selective, ATP-competitive P38 alpha MAP kinase inhibitors
To Be Published
|
|
5NGU
 
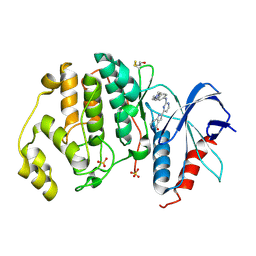 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
4YX2
 
 | | Crystal structure of Bovine prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
4YXL
 
 | | Crystal structure of Syrian hamster prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
4YXH
 
 | | Crystal structure of Deer prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
3MBI
 
 | | Crystal structure of the phosphoribosylpyrophosphate (PRPP) synthetase from Thermoplasma volcanium in complex with ADP-Mg2+ and ribose 5-phosphate | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
4YXK
 
 | | Crystal structure of Elk prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
3NAG
 
 | | Crystal structure of the phosphoribosylpyrophosphate (PRPP) synthetase from Thermoplasma Volcanium in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribose-phosphate pyrophosphokinase, ... | | Authors: | Cherney, M.M, Cherney, L.T, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
3GMU
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
4CO6
 
 | | Crystal structure of the Nipah virus RNA free nucleoprotein- phosphoprotein complex | | Descriptor: | BROMIDE ION, CHLORIDE ION, NUCLEOPROTEIN, ... | | Authors: | Yabukarksi, F, Lawrence, P, Tarbouriech, N, Bourhis, J.M, Jensen, M.R, Ruigrok, R.W.H, Blackledge, M, Volchkov, V, Jamin, M. | | Deposit date: | 2014-01-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure of Nipah Virus Unassembled Nucleoprotein in Complex with its Viral Chaperone.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3EJX
 
 | | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana in complex with LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3EKM
 
 | | Crystal structure of diaminopimelate epimerase form arabidopsis thaliana in complex with irreversible inhibitor DL-AziDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, van Belkum, M.J, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3FHZ
 
 | | Crystal structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and co-repressor, L-arginine | | Descriptor: | 5'-D(*TP*GP*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and Co-repressor, L-arginine.
J.Mol.Biol., 388, 2009
|
|
3EI6
 
 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-DAP: an external aldimine mimic | | Descriptor: | (2S,6S)-2-amino-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EIB
 
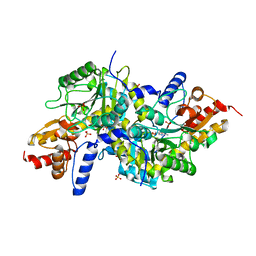 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
2D3Z
 
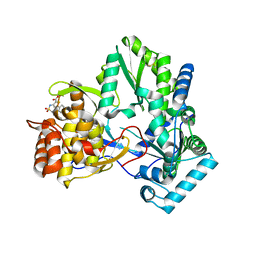 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3U
 
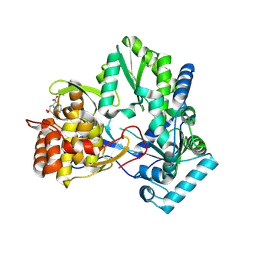 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
5TNC
 
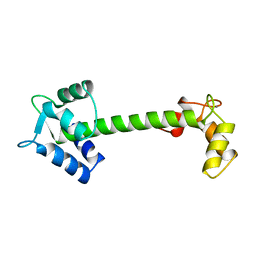 | |
