8RQH
 
 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
3DTM
 
 | |
8QQ3
 
 | | Streptavidin with a Ni-cofactor | | Descriptor: | 4-[4-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butylamino]-~{N}1,~{N}1'-di(quinolin-8-yl)cyclohexane-1,1-dicarboxamide, NICKEL (II) ION, Streptavidin | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial nickel chlorinase based on the biotin-streptavidin technology.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZMC
 
 | |
7ZMF
 
 | | Dehydratase domain of module 3 from Brevibacillus Brevis orphan BGC11 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMD
 
 | |
7ZMA
 
 | |
7ZM9
 
 | | Ketosynthase domain 3 of Brevibacillus Brevis orphan BGC11 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
4C16
 
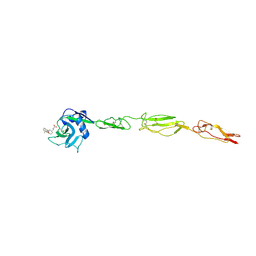 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | | Descriptor: | (1R,2R,3S)-3-methylcyclohexane-1,2-diol, (S)-CYCLOHEXYL LACTIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2013-08-09 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
6QGW
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody E6 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoE6, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
8B7S
 
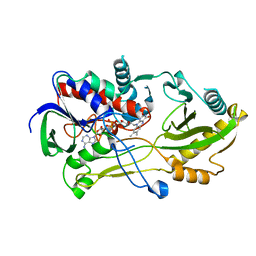 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
6SWR
 
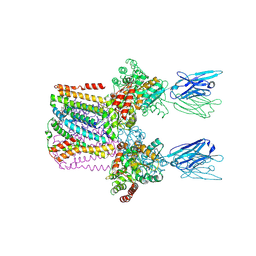 | | Crystal structure of the lysosomal potassium channel MtTMEM175 T38A mutant soaked with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody, Maltose/maltodextrin-binding periplasmic protein,Maltodextrin-binding protein,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2019-09-23 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K + channels.
Elife, 9, 2020
|
|
6QGX
 
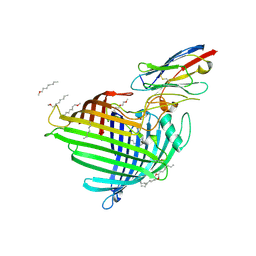 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
2X9B
 
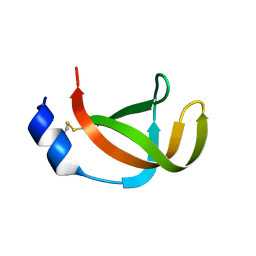 | | The filamentous phages fd and IF1 use different infection mechanisms | | Descriptor: | ATTACHMENT PROTEIN G3P | | Authors: | Lorenz, S.H, Jakob, R.P, Weininger, U, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
2X9A
 
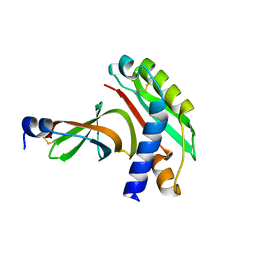 | | crystal structure of g3p from phage IF1 in complex with its coreceptor, the C-terminal domain of TolA | | Descriptor: | ATTACHMENT PROTEIN G3P, MEMBRANE SPANNING PROTEIN, REQUIRED FOR OUTER MEMBRANE INTEGRITY | | Authors: | Lorenz, S.H, Jakob, R.P, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
6ZPN
 
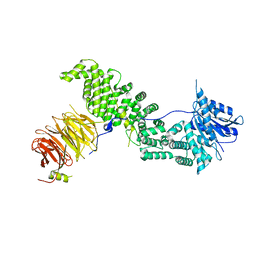 | | Crystal structure of Chaetomium thermophilum Raptor | | Descriptor: | WD_REPEATS_REGION domain-containing protein | | Authors: | Imseng, S, Boehm, R, Jakob, R.P, Hall, M.N, Hiller, S, Maier, T. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1.
Mol.Cell, 81, 2021
|
|
6HD8
 
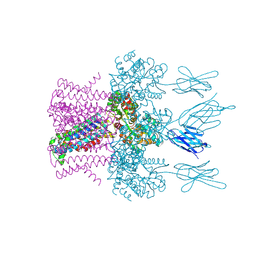 | | Crystal structure of the potassium channel MtTMEM175 in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HD9
 
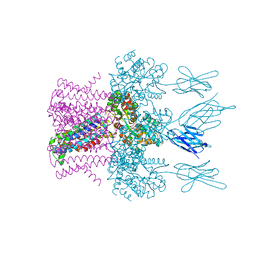 | | Crystal structure of the potassium channel MtTMEM175 with rubidium | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, RUBIDIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDA
 
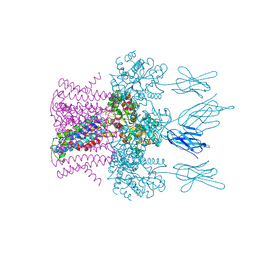 | | Crystal structure of the potassium channel MtTMEM175 with cesium | | Descriptor: | CESIUM ION, DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDC
 
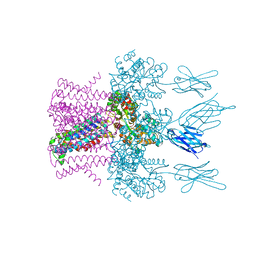 | | Crystal structure of the potassium channel MtTMEM175 T38A variant in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
8BP6
 
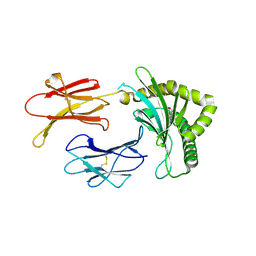 | | Structure of MHC-class I related molecule MR1 with bound M3Ade | | Descriptor: | (1R,5S)-8-(9H-purin-6-yl)-2-oxa-8-azabicyclo[3.3.1]nona-3,6-diene-4,6-dicarbaldehyde, Beta-2-microglobulin,Major histocompatibility complex class I-related gene protein | | Authors: | Berloffa, G, Jakob, R.P, Maier, T. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The carbonyl nucleoside adduct M3Ade stabilizes MR1 and activates MR1-restricted self- and tumor-reactive T cells
To Be Published
|
|
6HDB
 
 | | Crystal structure of the potassium channel MtTMEM175 with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
8AS2
 
 | | Structure of arrestin2 in complex with 4P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
