3V3M
 
 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
4UZW
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4UZX
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1H1J
 
 | |
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
3BM9
 
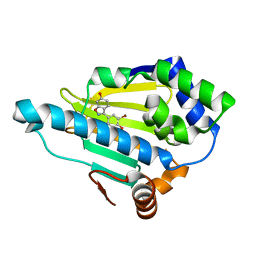 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-bromo-6-(6-hydroxy-1,2-benzisoxazol-3-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
3BMY
 
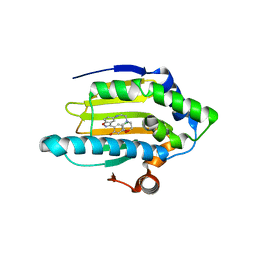 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-chloro-6-{5-[(2-morpholin-4-ylethyl)amino]-1,2-benzisoxazol-3-yl}benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
4WEB
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMAMIDE, Mouse Fab Heavy Chain, ... | | Authors: | Khan, A.G, Whidby, J, Miller, M.T, Scarborough, H, Zatorski, A.V, Cygan, A, Price, A.A, Yost, S.A, Bohannon, C.D, Jacob, J, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2.
Nature, 509, 2014
|
|
3BEW
 
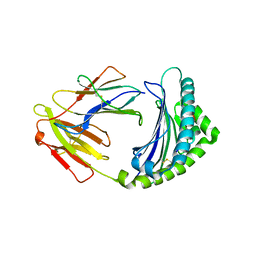 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
2HLI
 
 | | Solution structure of Crotonaldehyde-Derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct in the 5'-CpG-3' Sequence | | Descriptor: | DNA dodecamer, DNA dodecamer with S-crotonaldehyde adduct | | Authors: | Cho, Y.-J, Wang, H, Kozekov, I.D, Kurtz, A.J, Jacob, J, Voehler, M, Smith, J, Harris, T.M, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Orientation of the Crotonaldehyde-Derived N(2)-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct Hinders Interstrand Cross-Link Formation in the 5'-CpG-3' Sequence
Chem.Res.Toxicol., 19, 2006
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
1HHW
 
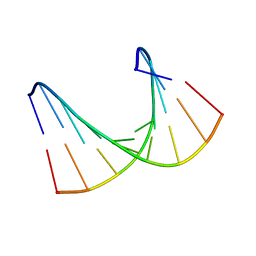 | | Solution structure of LNA1:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*+TLNP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1HHX
 
 | | Solution structure of LNA3:RNA hybrid | | Descriptor: | 5- D(*CP*+TP*GP*AP*+TP*AP*+TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
214D
 
 | | THE SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING A SINGLE 2'-O-METHYL-BETA-D-ARAT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*AP*TP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*AP*(T41)P*AP*TP*GP*CP*G)-3') | | Authors: | Gotfredsen, C.H, Spielmann, H.P, Wengel, J, Jacobsen, J.P. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA duplex containing a single 2'-O-methyl-beta-D-araT: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Bioconjug.Chem., 7, 1996
|
|
2KDY
 
 | | NMR structure of LP2086-B01 | | Descriptor: | Factor H binding protein variant B01_001 | | Authors: | Mascioni, A, Bentley, B.E, Camarda, R, Dilts, D.A, Fink, P, Gusarova, V, Hoiseth, S, Jacob, J, Lin, S.L, Malakian, K, McNeil, L.K, Mininni, T, Moy, F, Murphy, E, Novikova, E, Sigethy, S, Wen, Y, Zlotnick, G.W, Tsao, D.H.H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
J.Biol.Chem., 284, 2009
|
|
1D8V
 
 | | THE RESTRAINED AND MINIMIZED AVERAGE NMR STRUCTURE OF MAP30. | | Descriptor: | ANTI-HIV AND ANTI-TUMOR PROTEIN MAP30 | | Authors: | Wang, Y.-X, Neamati, N, Jacob, J, Palmer, I, Stahl, S.J. | | Deposit date: | 1999-10-26 | | Release date: | 1999-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of anti-HIV-1 and anti-tumor protein MAP30: structural insights into its multiple functions.
Cell(Cambridge,Mass.), 99, 1999
|
|
4AUC
 
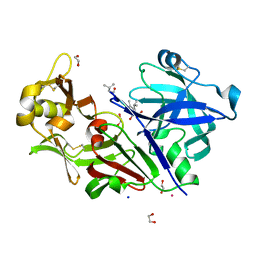 | | Bovine chymosin in complex with Pepstatin A | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHYMOSIN, ... | | Authors: | Langholm Jensen, J, Navarro Poulsen, J.C, Jacobsen, J, van den Brink, J.M, Qvist, K.B, Larsen, S. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of Bovine Chymosin in Complex with Pepstatin A
To be Published
|
|
108D
 
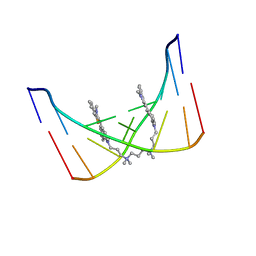 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
4M1U
 
 | | The crystal structure of Stx2 and a disaccharide ligand | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-methyl beta-D-galactopyranoside, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 A-subunit, ... | | Authors: | Yin, J, James, M.N.G, Jacobson, J.M, Kitov, P.I, Bundle, D.R, Mulvey, G, Armstrong, G. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The crystal structure of shiga toxin type 2 with bound disaccharide guides the design of a heterobifunctional toxin inhibitor.
J.Biol.Chem., 289, 2014
|
|
1OCI
 
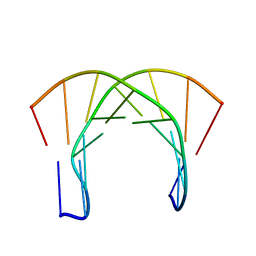 | | [3.2.0]bcANA:DNA | | Descriptor: | 5'-D(*CP*TP*GP*A TLBP*AP*TP*GP*CP)-3', 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*GP)-3' | | Authors: | Tommerholt, H.V, Christensen, N.K, Nielsen, P, Wengel, J, Stein, P.C, Jacobsen, J.P, Petersen, M. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a DsDNA Containing a Bicyclic D-Arabino-Configured Nucleotide Fixed in an O4'-Endo Sugar Conformation
Org.Biomol.Chem., 1, 2003
|
|
1H0Q
 
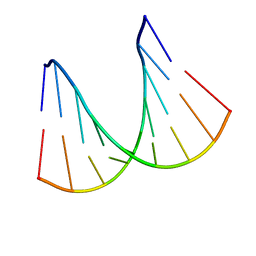 | | NMR solution structure of a fully modified locked nucleic acid (LNA) hybridized to RNA | | Descriptor: | 5-D(*(LKC)P*(TLN)P*(LCG)P*(LCA)P*(TLN)P*(LCA)P* (TLN)P*(LCG)P*(LCC))-3, 5-R(*GP*CP*AP*UP*AP*UP*CP*AP*G)-3 | | Authors: | Rasmussen, J, Petersen, M, Nielsen, K.E, Kumar, R, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2002-06-27 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of Fully Modified Locked Nucleic Acid (Lna) Hybrids: Solution Structure of an Lna:RNA Hybrid and Characterization of an Lna:RNA Hybrid
Bioconjug.Chem., 15, 2004
|
|
1HG9
 
 | | Solution structure of DNA:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*TP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-13 | | Release date: | 2002-01-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity.
Chemistry, 6, 2000
|
|
1GV6
 
 | | Solution structure of alfa-L-LNA:DNA duplex | | Descriptor: | 5- D(*CP*(ATL)P*GP*CP*(ATL)P*(ATL)P*CP*(ATL)P* GP*C) -3, 5- D(*GP*CP*AP*GP*AP*AP*GP*CP*AP*G) -3 | | Authors: | Nielsen, K.M.E, Petersen, M, Haakansson, A.E, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alfa-L-Lna (Alfa-L-Ribo Configured Locked Nucleic Acid) Recognition of DNA: An NMR Spectroscopic Study
Chemistry, 8, 2002
|
|
