1MYW
 
 | | CRYSTAL STRUCTURE OF A YELLOW FLUORESCENT PROTEIN WITH IMPROVED MATURATION AND REDUCED ENVIRONMENTAL SENSITIVITY | | Descriptor: | Green fluorescent protein | | Authors: | Rekas, A, Alattia, J.R, Nagai, T, Miyawaki, A, Ikura, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Venus, a Yellow Fluorescent
Protein with Improved Maturation and
Reduced Environmental Sensitivity
J.Biol.Chem., 277, 2002
|
|
1MR3
 
 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | Authors: | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
7LVT
 
 | | Structure of full-length GluK1 with L-Glu | | Descriptor: | Isoform Glur5-2 of Glutamate receptor ionotropic, kainate 1 | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural and compositional diversity in the kainate receptor family.
Cell Rep, 37, 2021
|
|
7LJH
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Zn+2 bound | | Descriptor: | Poly(Aspartic acid) hydrolase, ZINC ION | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
7LJI
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Gd+3 bound | | Descriptor: | GADOLINIUM ION, Poly(Aspartic acid) hydrolase | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
5CYG
 
 | |
4UW8
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
5L3H
 
 | |
5MIL
 
 | | Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity islands transfer. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase family protein, MAGNESIUM ION | | Authors: | Donderis, J, Marina, A, Penades, J.R. | | Deposit date: | 2016-11-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity island transfer.
Elife, 6, 2017
|
|
7QEL
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247 | | Descriptor: | (2~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)spiro[2.3]hexane-2-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247
To Be Published
|
|
7RNV
 
 | |
7RNS
 
 | |
7RNU
 
 | |
7RFK
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor Sinefungin | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFM
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFL
 
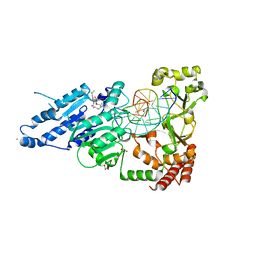 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC0946 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFN
 
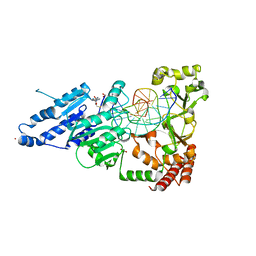 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC8158 | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RRO
 
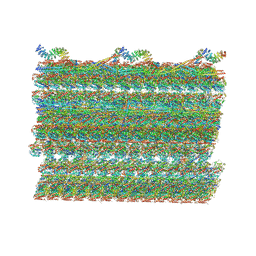 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
5DJ0
 
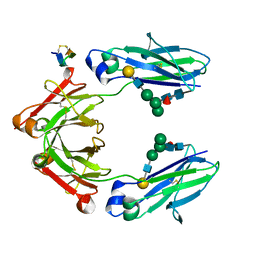 | | Fc Heterodimer Design 11.2 Y349S/K370Y + E357D/S364Q | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5DJ2
 
 | | Fc Heterodimer Design 7.4 Y407A + T366V/K409V | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Secrist, E, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5DJD
 
 | | Fc Heterodimer Design 5.1 T366V + Y407F | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-02 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5CTA
 
 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 10% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5DBL
 
 | |
