4QHI
 
 | | Crystal structure of Methanocaldococcus jannaschii selecase mutant R36W | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4Q6I
 
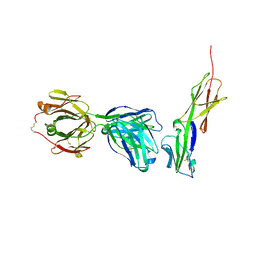 | | Crystal structure of murine 2D5 Fab, a potent anti-CD4 HIV-1-neutralizing antibody in complex with CD4 | | Descriptor: | Heavy chain of murine 2D5 Fab, Light chain of murine 2D5 Fab, T-cell surface glycoprotein CD4 | | Authors: | Boyington, J.C, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor.
Sci Transl Med, 6, 2014
|
|
4QLK
 
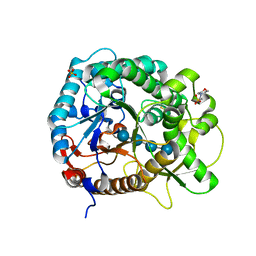 | | Crystal structure of rice BGlu1 E176Q/Y341A mutant complexed with cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2014-06-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Effects of active site cleft residues on oligosaccharide binding, hydrolysis, and glycosynthase activities of rice BGlu1 and its mutants
Protein Sci., 23, 2014
|
|
4QHJ
 
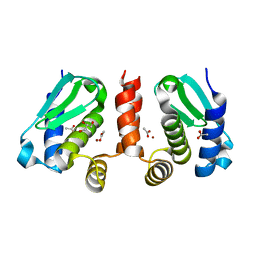 | | Crystal structure of Methanocaldococcus jannaschii selecase mutant I100F+H107F | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6SX1
 
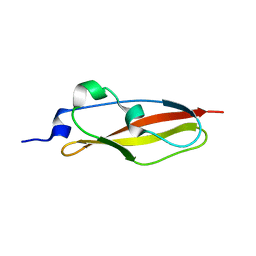 | | Structure of Rib Standard, a Rib domain from Lactobacillus acidophilus | | Descriptor: | Surface protein | | Authors: | Cooper, R.E.M, Griffiths, S.C, Turkenburg, J.P, Bateman, A, Potts, J.R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SE1
 
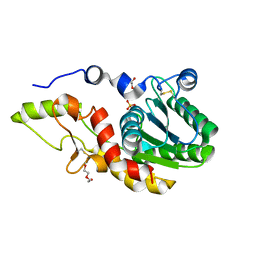 | | Structure of Salmonella ser. Paratyphi A lipopolysaccharide acetyltransferase periplasmic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative lipopolysaccharide modification acyltransferase, SULFATE ION | | Authors: | Tindall, S.N, Pearson, C, Herman, R, Jenkins, H.T, Thomas, G.H, Potts, J.R, Van der Woude, M. | | Deposit date: | 2019-07-29 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Acetylation of Surface Carbohydrates in Bacterial Pathogens Requires Coordinated Action of a Two-Domain Membrane-Bound Acyltransferase.
Mbio, 11, 2020
|
|
6YGN
 
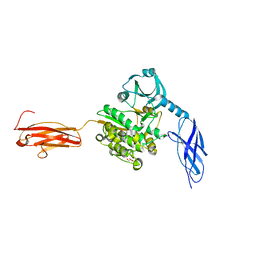 | | Titin kinase and its flanking domains | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Titin | | Authors: | Fleming, J.R, Franke, B, Bogomolovas, J, Mayans, O. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Titin kinase ubiquitination aligns autophagy receptors with mechanical signals in the sarcomere.
Embo Rep., 22, 2021
|
|
4PYZ
 
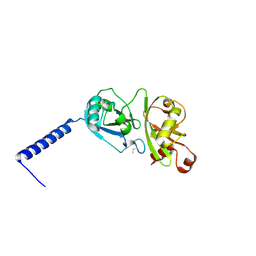 | | Crystal structure of the first two Ubl domains of Deubiquitylase USP7 | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Walker, J.R, Dong, A, Ong, M.S, Dhe-Paganon, S, Kania, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the first two Ubl domains of Deubiquitylase USP7
to be published
|
|
7AYY
 
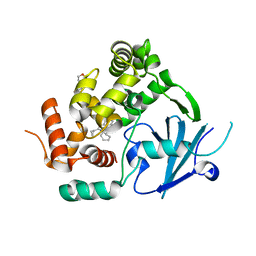 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
 | |
7AZ0
 
 | |
4Q2W
 
 | | Crystal Structure of pneumococcal peptidoglycan hydrolase LytB | | Descriptor: | GLYCEROL, Putative endo-beta-N-acetylglucosaminidase | | Authors: | Bai, X.H, Chen, H.J, Jiang, Y.L, Wen, Z, Cheng, W, Li, Q, Zhang, J.R, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis.
J.Biol.Chem., 289, 2014
|
|
4Q84
 
 | | Apo YcaO | | Descriptor: | MERCURY (II) ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4QHG
 
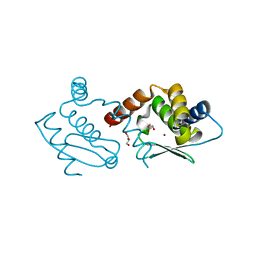 | | Crystal structure of Methanocaldococcus jannaschii dimeric selecase | | Descriptor: | GLYCEROL, Uncharacterized protein MJ1213, ZINC ION | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6SIH
 
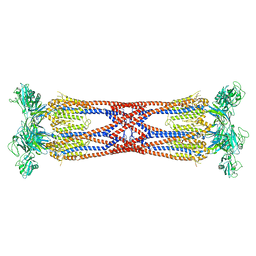 | |
4QLJ
 
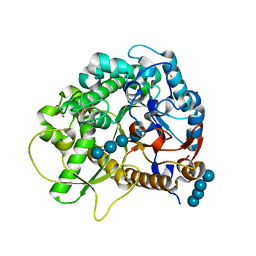 | | Crystal structure of rice BGlu1 E386G/Y341A/Q187A mutant complexed with cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2014-06-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effects of active site cleft residues on oligosaccharide binding, hydrolysis, and glycosynthase activities of rice BGlu1 and its mutants
Protein Sci., 23, 2014
|
|
7BZM
 
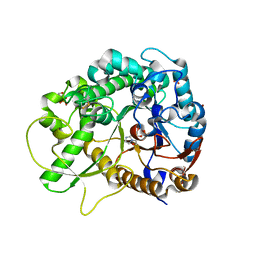 | | Crystal structure of rice Os3BGlu7 with glucoimidazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLUCOIMIDAZOLE, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R, Tankrathok, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7.
Biomolecules, 10, 2020
|
|
4QHH
 
 | | Crystal structure of Methanocaldococcus jannaschii tetrameric selecase | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4P16
 
 | | Crystal structure of the papain-like protease of Middle-East Respiratory Syndrome coronavirus | | Descriptor: | ORF1a, ZINC ION | | Authors: | Lei, J, Mesters, J.R, Ma, Q, Hilgenfeld, R. | | Deposit date: | 2014-02-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features.
Antiviral Res., 109C, 2014
|
|
4PBB
 
 | |
6Z8F
 
 | | Human Picobirnavirus D45-CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6Z8D
 
 | | Human Picobirnavirus CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
4PLH
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with oxamate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, OXAMIC ACID, SODIUM ION, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
6Z8E
 
 | | Human Picobirnavirus Ht-CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6ZVN
 
 | |
