2QWD
 
 | |
2G9E
 
 | | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TRAM | | Descriptor: | Protein traM | | Authors: | Lu, J, Edwards, R.A, Wong, J.J, Manchak, J, Scott, P.G, Frost, L.S, Glover, J.N. | | Deposit date: | 2006-03-06 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM.
Embo J., 25, 2006
|
|
2GKI
 
 | | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity | | Descriptor: | nuclease | | Authors: | Kim, Y.R, Kim, J.S, Lee, S.H, Lee, W.R, Sohn, J.N, Chung, Y.C, Shim, H.K, Lee, S.C, Kwon, M.H, Kim, Y.S. | | Deposit date: | 2006-04-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity.
J.Biol.Chem., 281, 2006
|
|
1SRA
 
 | | STRUCTURE OF A NOVEL EXTRACELLULAR CA2+-BINDING MODULE IN BM-40(SLASH)SPARC(SLASH)OSTEONECTIN | | Descriptor: | CALCIUM ION, SPARC | | Authors: | Hohenester, E, Maurer, P, Hohenadl, C, Timpl, R, Jansonius, J.N, Engel, J. | | Deposit date: | 1995-08-21 | | Release date: | 1996-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a novel extracellular Ca(2+)-binding module in BM-40.
Nat.Struct.Biol., 3, 1996
|
|
1BTV
 
 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
4XW3
 
 | |
1U20
 
 | |
1CJ0
 
 | | CRYSTAL STRUCTURE OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE AT 2.8 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (SERINE HYDROXYMETHYLTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Scarsdale, J.N, Kazanina, G, Radaev, S, Schirch, V, Wright, H.T. | | Deposit date: | 1999-04-20 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rabbit cytosolic serine hydroxymethyltransferase at 2.8 A resolution: mechanistic implications.
Biochemistry, 38, 1999
|
|
4Y76
 
 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y79
 
 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
3OA5
 
 | | The structure of chi1, a chitinase from Yersinia entomophaga | | Descriptor: | Chi1, GLYCEROL, NONAETHYLENE GLYCOL | | Authors: | Busby, J.N, Lott, J.S, Hurst, M.R.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of Chi1 Chitinase from Yen-Tc: the multisubunit insecticidal ABC toxin complex of Yersinia entomophaga
J.Mol.Biol., 415, 2012
|
|
1DFO
 
 | | CRYSTAL STRUCTURE AT 2.4 ANGSTROM RESOLUTION OF E. COLI SERINE HYDROXYMETHYLTRANSFERASE IN COMPLEX WITH GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Scarsdale, J.N, Radaev, S, Kazanina, G, Schirch, V, Wright, H.T. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate.
J.Mol.Biol., 296, 2000
|
|
3P4P
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with fumarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andr ll, J, Luna-Ch vez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
1W8N
 
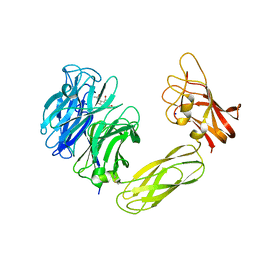 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1W8O
 
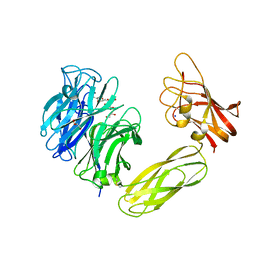 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens | | Descriptor: | BACTERIAL SIALIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
3P4Q
 
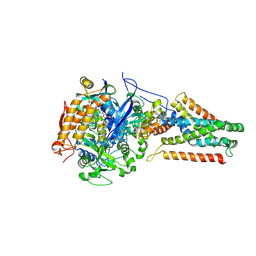 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4R
 
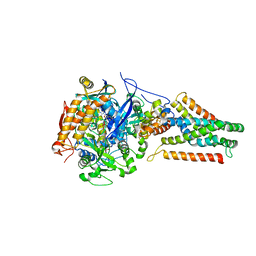 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with glutarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
1DY3
 
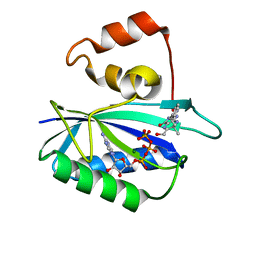 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
3P4S
 
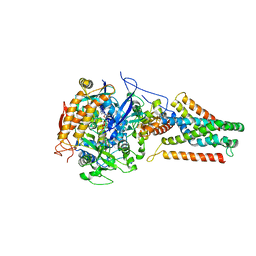 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with a 3-nitropropionate adduct | | Descriptor: | 3-NITROPROPANOIC ACID, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
4YE7
 
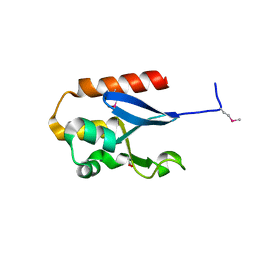 | |
1V2I
 
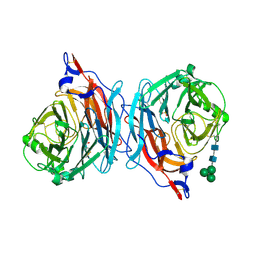 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1V3B
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1V3C
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1T66
 
 | | The structure of FAB with intermediate affinity for fluorescein. | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, immunoglobulin heavy chain, immunoglobulin light chain | | Authors: | Terzyan, S, Ramsland, P.A, Voss Jr, E.W, Herron, J.N, Edmundson, A.B. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional Structures of Idiotypically Related Fabs with Intermediate and High Affinity for Fluorescein.
J.Mol.Biol., 339, 2004
|
|
1V3E
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with ZANAMAVIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
