3MCG
 
 | |
2BTV
 
 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | Descriptor: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | Authors: | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 1998-09-05 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
1CBV
 
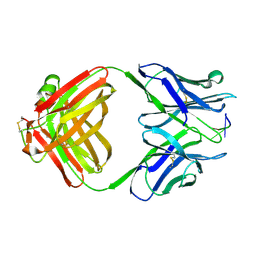 | | AN AUTOANTIBODY TO SINGLE-STRANDED DNA: COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF THE UNLIGANDED FAB AND A DEOXYNUCLEOTIDE-FAB COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (FAB (BV04-01) AUTOANTIBODY-HEAVY CHAIN), PROTEIN (FAB (BV04-01) AUTOANTIBODY-LIGHT CHAIN) | | Authors: | Herron, J.N, He, X.M, Ballard, D.W, Blier, P.R, Pace, P.E, Bothwell, A.L.M, Voss Junior, E.W, Edmundson, A.B. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | An autoantibody to single-stranded DNA: comparison of the three-dimensional structures of the unliganded Fab and a deoxynucleotide-Fab complex.
Proteins, 11, 1991
|
|
4BP7
 
 | | Asymmetric structure of a virus-receptor complex | | Descriptor: | COAT PROTEIN | | Authors: | Dent, K.C, Thompson, R, Barker, A.M, Barr, J.N, Hiscox, J.A, Stockley, P.G, Ranson, N.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (39 Å) | | Cite: | The Asymmetric Structure of an Icosahedral Virus Bound its Receptor Suggests a Mechanism for Genome Release.
Structure, 21, 2013
|
|
8OVH
 
 | |
4V5M
 
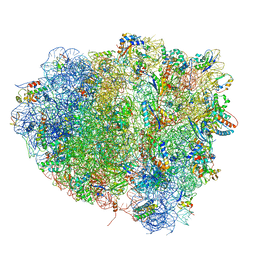 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
2SBT
 
 | | A COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF SUBTILISIN BPN AND SUBTILISIN NOVO | | Descriptor: | ACETONE, SUBTILISIN NOVO | | Authors: | Drenth, J, Hol, W.G.J, Jansonius, J.N, Koekoek, R. | | Deposit date: | 1976-09-07 | | Release date: | 1976-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo.
Cold Spring Harbor Symp.Quant.Biol., 36, 1972
|
|
1SJ2
 
 | | Crystal structure of Mycobacterium tuberculosis catalase-peroxidase | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase/catalase T | | Authors: | Bertrand, T, Eady, N.A.J, Jones, J.N, Bodiguel, J, Jesmin, Nagy, J.M, Raven, E.L, Jamart-Gregoire, B, Brown, K.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Catalase-Peroxidase.
J.Biol.Chem., 279, 2004
|
|
6KL5
 
 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL6
 
 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
4V5N
 
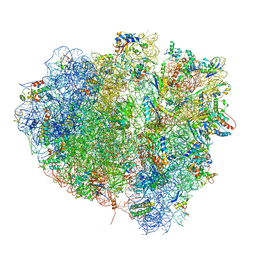 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
7NN9
 
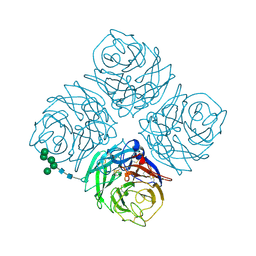 | | NATIVE INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Varghese, J.N, Colman, P.M. | | Deposit date: | 1995-03-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase.
Protein Sci., 4, 1995
|
|
7S3T
 
 | | NzeB Diketopiperazine Dimerase Mutant: Q68I-G87A-A89G-I90V | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Harris, N.R, Shende, V.V, Sanders, J.N, Newmister, S.A, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 2023
|
|
5ZWB
 
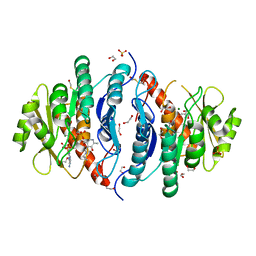 | | Crystal structure of Pyridoxal kinase (PdxK) from Salmonella typhimurium in complex with ADP, PL-linked to Lys233 via a Schiff base | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Deka, G, Benazir, J.F, Kalyani, J.N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional studies on Salmonella typhimurium pyridoxal kinase: the first structural evidence for the formation of Schiff base with the substrate.
Febs J., 286, 2019
|
|
5ZW9
 
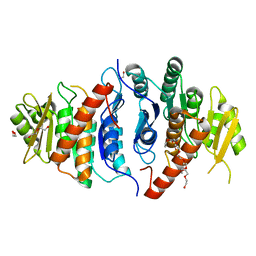 | | Crystal structure of Pyridoxal kinase (PdxK) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Deka, G, Benazir, J.F, Kalyani, J.N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies on Salmonella typhimurium pyridoxal kinase: the first structural evidence for the formation of Schiff base with the substrate.
Febs J., 286, 2019
|
|
7S3J
 
 | | Crystal Structure of AspB P450 in complex with brevianamide F substrates | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, AspB, GLYCEROL, ... | | Authors: | Newmister, S.A, Shende, V.V, Harris, N.R, Sanders, J.N, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-07 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5ZWA
 
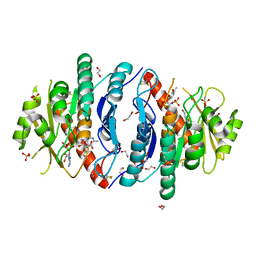 | | Crystal structure of Pyridoxal kinase (PdxK) from Salmonella typhimurium in complex with ADP, PL-linked to Lys233 via Schiff base in protomer A and the product (PLP) in protomer B | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Deka, G, Benazir, J.F, Kalyani, J.N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies on Salmonella typhimurium pyridoxal kinase: the first structural evidence for the formation of Schiff base with the substrate.
Febs J., 286, 2019
|
|
7AOY
 
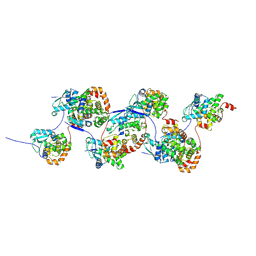 | |
7DYD
 
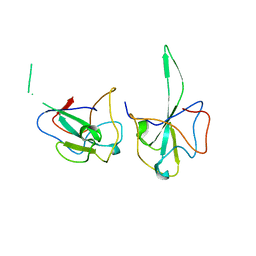 | |
8CCX
 
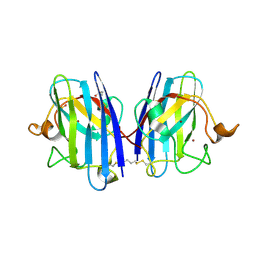 | | Human SOD1 in complex with S-XL6 cross-linker | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-01-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
7AAT
 
 | |
6NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
8Q6M
 
 | | Human SOD1 low dose data collecton | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-08-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
5JP3
 
 | |
5JP1
 
 | | Structure of Xanthomonas campestris effector protein XopD bound to tomato SUMO | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, MALONATE ION, Small ubiquitin-related modifier, ... | | Authors: | Pruneda, J.N, Komander, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases.
Mol.Cell, 63, 2016
|
|
