5NTJ
 
 | |
5O20
 
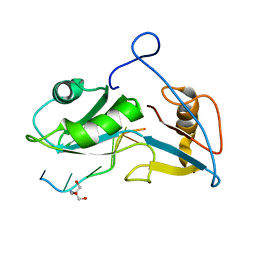 | | Structure of Nrd1 RNA binding domain in complex with RNA (UUAGUAAUCC) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, RNA (5'-R(*UP*AP*GP*UP*AP*AP*UP*C)-3') | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O9L
 
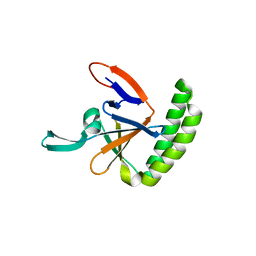 | | Crystal structure of human Histamine-Releasing Factor (HRF/TCTP) | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
2NV5
 
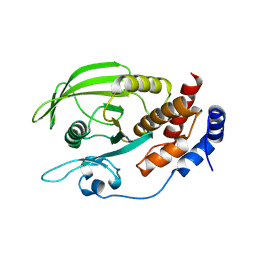 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | PTPRD, PHOSPHATASE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2NX2
 
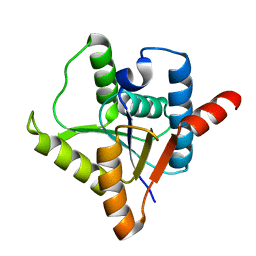 | | Crystal structure of protein ypsA from Bacillus subtilis, Pfam DUF1273 | | Descriptor: | Hypothetical protein ypsA | | Authors: | Ramagopal, U.A, Alvarado, J, Dickey, M, Reyes, C, Toro, R, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein ypsA from Bacillus subtilis.
To be Published
|
|
2NS9
 
 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
2O3A
 
 | | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus | | Descriptor: | UPF0106 protein AF_0751 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Slocombe, A, Sridhar, V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus
To be Published
|
|
2NXA
 
 | |
2O16
 
 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
2O5K
 
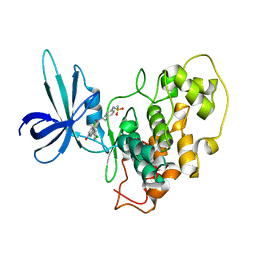 | | Crystal Structure of GSK3beta in complex with a benzoimidazol inhibitor | | Descriptor: | 2-(2,4-DICHLORO-PHENYL)-7-HYDROXY-1H-BENZOIMIDAZOLE-4-CARBOXYLIC ACID [2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-AMIDE, Glycogen synthase kinase-3 beta | | Authors: | Shin, D, Lee, S.C, Heo, Y.S, Cho, Y.S, Kim, Y.E, Hyun, Y.L, Cho, J.M, Lee, Y.S, Ro, S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OXL
 
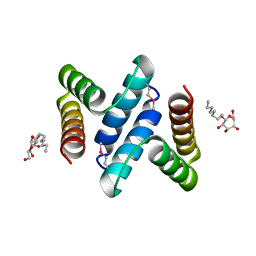 | | Structure and Function of the E. coli Protein YmgB: a Protein Critical for Biofilm Formation and Acid Resistance | | Descriptor: | Hypothetical protein ymgB, octyl beta-D-glucopyranoside | | Authors: | Page, R, Peti, W, Woods, T.K, Palermino, J.M, Doshi, O. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance.
J.Mol.Biol., 373, 2007
|
|
5NWA
 
 | |
2OHW
 
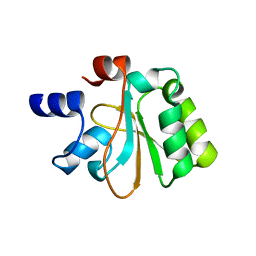 | | Crystal structure of the YueI protein from Bacillus subtilis | | Descriptor: | YueI protein | | Authors: | Bonanno, J.B, Jin, X, Mu, H, Dickey, M, Bain, K.T, Wu, B, Chen, T, Reyes, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-10 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the YueI protein from Bacillus subtilis
To be Published
|
|
2NR4
 
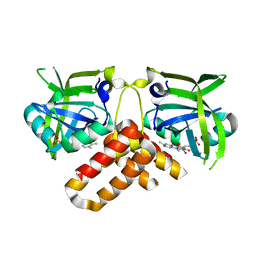 | | Crystal structure of FMN-bound protein MM1853 from Methanosarcina mazei, Pfam DUF447 | | Descriptor: | Conserved hypothetical protein, FLAVIN MONONUCLEOTIDE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved FMN bound hypothetical protein from Methanosarcina mazei
To be Published
|
|
2NZJ
 
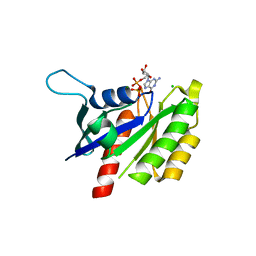 | | The crystal structure of REM1 in complex with GDP | | Descriptor: | CHLORIDE ION, GTP-binding protein REM 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turnbull, A.P, Papagrigoriou, E, Ugochukwu, E, Elkins, J.M, Soundararajan, M, Yang, X, Gorrec, F, Umeano, C, Salah, E, Burgess, N, Johansson, C, Berridge, G, Gileadi, O, Bray, J, Marsden, B, Watts, S, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-23 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of REM1 in complex with GDP
To be Published
|
|
5O0E
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp120 and Fasudil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, RKp120, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-30 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
2ODV
 
 | |
5O2D
 
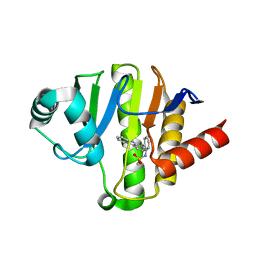 | | PARP14 Macrodomain 2 with inhibitor | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}-[2-(9~{H}-carbazol-1-yl)phenyl]methanesulfonamide | | Authors: | Uth, K, Schuller, M, Sieg, C, Wang, J, Krojer, T, Knapp, S, Riedels, K, Bracher, F, Edwards, A.M, Arrowsmith, C, Bountra, C, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
ACS Chem. Biol., 12, 2017
|
|
5O1Y
 
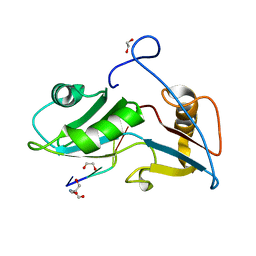 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
2NZ1
 
 | |
5OHQ
 
 | |
5OIK
 
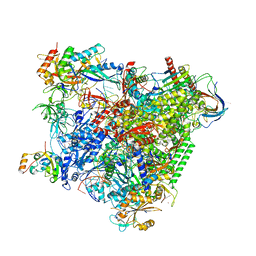 | | Structure of an RNA polymerase II-DSIF transcription elongation complex | | Descriptor: | DNA (43-MER), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Bernecky, C, Plitzko, J.M, Cramer, P. | | Deposit date: | 2017-07-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2NYP
 
 | |
2NYZ
 
 | |
2NZF
 
 | |
