2UXE
 
 | | The structure of Vaccinia virus N1 | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Cooray, S, Bahar, M.W, Abrescia, N.G.A, McVey, C.E, Bartlett, N.W, Chen, R.A.-J, Stuart, D.I, Grimes, J.M, Smith, G.L. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and Structural Studies of the Vaccinia Virus Virulence Factor N1 Reveal a Bcl-2-Like Anti- Apoptotic Protein
J.Gen.Virol., 88, 2007
|
|
4HUP
 
 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)-L-phenylalanyl)-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-3-phenyl-propanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
2V12
 
 | | Crystal Structure of Renin with Inhibitor 8 | | Descriptor: | N-[(2S,4S,5S,7R)-4-AMINO-8-(BUTYLAMINO)-5-HYDROXY-7-METHYL-2-(1-METHYLETHYL)-8-OXOOCTYL]-2-(3-METHOXYPROPOXY)BENZAMIDE, RENIN | | Authors: | Rahuel, J, Rasetti, V, Maibaum, J, Rueger, H, Goschke, R, Cohen, N.C, Stutz, S, Cumin, F, Fuhrer, W, Wood, J.M, Grutter, M.G. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Based Drug Design: The Discovery of Novel Nonpeptide Orally Active Inhibitors of Human Renin
Chem.Biol., 7, 2000
|
|
2V3Z
 
 | |
4HV7
 
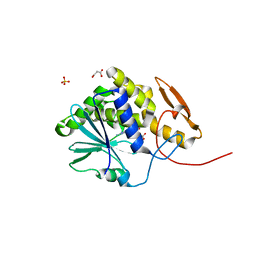 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)glycine | | Descriptor: | 2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]ethanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-05 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
6FD2
 
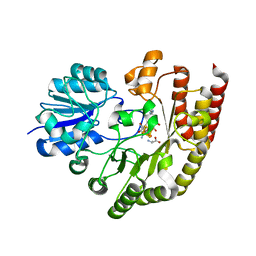 | | Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Liu, W.Q, Amara, P, Mouesca, J.M, Ji, X, Renoux, O, Martin, L, Zhang, C, Zhang, Q, Nicolet, Y. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility.
J. Am. Chem. Soc., 140, 2018
|
|
2VHC
 
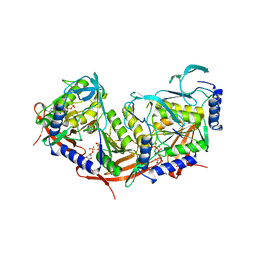 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
6F62
 
 | |
2V3X
 
 | |
2V1Y
 
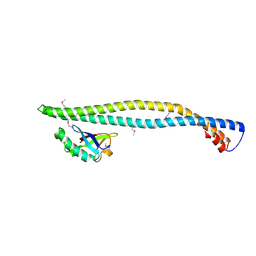 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
6EVS
 
 | | Characterization of 2-deoxyribosyltransferase from psychrotolerant bacterium Bacillus psychrosaccharolyticus: a suitable biocatalyst for the industrial synthesis of antiviral and antitumoral nucleosides | | Descriptor: | N-deoxyribosyltransferase | | Authors: | Fresco-Tabohada, A, Fernandez-Lucas, J, Acebal, C, Arroyo, M, Ramon, F, Mancheno, J.M, de la Mata, I. | | Deposit date: | 2017-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2'-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium
Catalysts, 2019
|
|
2W3Z
 
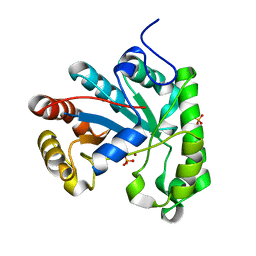 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
4HJK
 
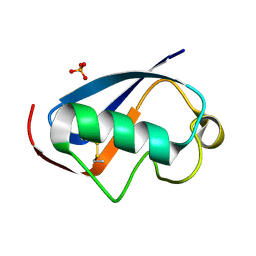 | | U7Ub7 Disulfide variant | | Descriptor: | PHOSPHATE ION, Ubiquitin | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2012-10-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Potent and selective inhibitors of USP7/HAUSP activity by protein conformational stabilization
to be published
|
|
2WLZ
 
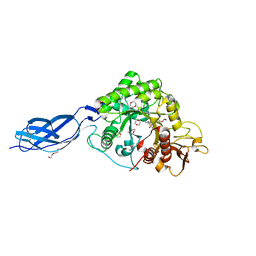 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6F5O
 
 | |
2WO8
 
 | | MMP12 complex with a beta hydroxy carboxylic acid | | Descriptor: | (3S)-5-biphenyl-4-yl-3-hydroxypentanoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Holmes, I.P, Gaines, S, Watson, S.P, Lorthioir, O, Walker, A, Baddeley, S.J, Herbert, S, Egan, D, Convery, M.A, Singh, O.M.P, Gross, J.W, Strelow, J.M, Smith, R.H, Amour, A.J, Brown, D, Martin, S.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2X6A
 
 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, PHOSPHOCHOLINE, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
6EYO
 
 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
4I1K
 
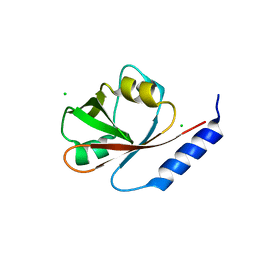 | | Crystal Structure of VRN1 (Residues 208-341) | | Descriptor: | B3 domain-containing transcription factor VRN1, CHLORIDE ION | | Authors: | King, G, Chanson, A.H, McCallum, E.J, Ohme-Takagi, M, Byriel, K, Hill, J.M, Martin, J.L, Mylne, J.S. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface.
J.Biol.Chem., 288, 2013
|
|
6F64
 
 | | Crystal structure of the SYCP1 C-terminal back-to-back assembly | | Descriptor: | ACETATE ION, Synaptonemal complex protein 1 | | Authors: | Dunce, J.M, Millan, C, Uson, I, Davies, O.R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-06-06 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2WC4
 
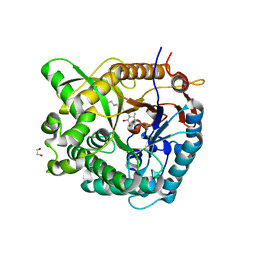 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-thia-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
6EYN
 
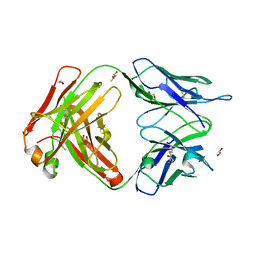 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
2WD1
 
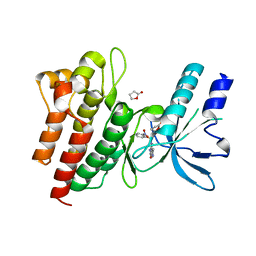 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6F5P
 
 | |
2W0U
 
 | | CRYSTAL STRUCTURE OF HUMAN GLYCOLATE OXIDASE IN COMPLEX WITH THE INHIBITOR 5-[(4-CHLOROPHENYL)SULFANYL]- 1,2,3-THIADIAZOLE-4-CARBOXYLATE. | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, HYDROXYACID OXIDASE 1 | | Authors: | Bourhis, J.M, Lindqvist, Y. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Human Glycolate Oxidase in Complex with the Inhibitor 4-Carboxy-5-[(4-Chlorophenyl)Sulfanyl]-1,2,3-Thiadiazole.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
