4FAL
 
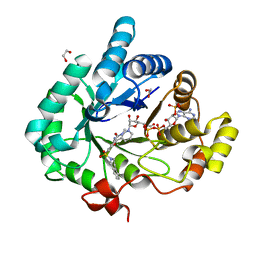 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)-N-methylbenzamide (80) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)-N-methylbenzamide, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
4FA3
 
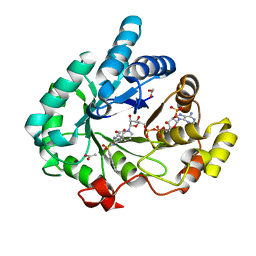 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid (86) | | Descriptor: | (3R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
2CYG
 
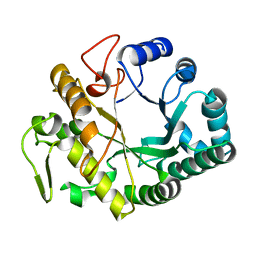 | | Crystal structure at 1.45- resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome | | Descriptor: | beta-1, 3-glucananse | | Authors: | Receveur-Brechot, V, Czjzek, M, Barre, A, Roussel, A, Peumans, W.J, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45-A resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome
Proteins, 63, 2006
|
|
4GB1
 
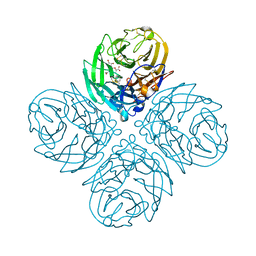 | | Synthesis and Evaluation of Novel 3-C-alkylated-Neu5Ac2en Derivatives as Probes of Influenza Virus Sialidase 150-loop flexibility | | Descriptor: | 5-acetamido-2,6-anhydro-3,5-dideoxy-3-[(2E)-3-phenylprop-2-en-1-yl]-D-glycero-L-altro-non-2-enonic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Rudrawar, S, Rameix-Welti, M.-A, Maggioni, A, Dyason, J.C, Rose, F.J, van der Werf, S, Thomson, R.J, Naffakh, N, Russell, R.J.M, von Itzstein, M. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Synthesis and evaluation of novel 3-C-alkylated-Neu5Ac2en derivatives as probes of influenza virus sialidase 150-loop flexibility.
Org.Biomol.Chem., 10, 2012
|
|
6AFH
 
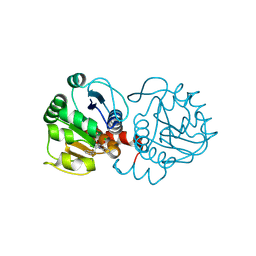 | | DJ-1 with compound 10 | | Descriptor: | 1-(2-phenylethyl)indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFA
 
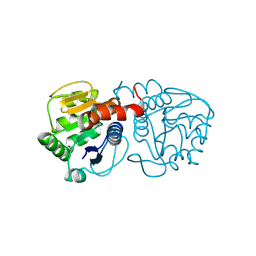 | | DJ-1 with isatin (low concentration) | | Descriptor: | CHLORIDE ION, ISATIN, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFE
 
 | | DJ-1 with compound 7 | | Descriptor: | 7-(trifluoromethyl)-1~{H}-indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AF9
 
 | |
6AFF
 
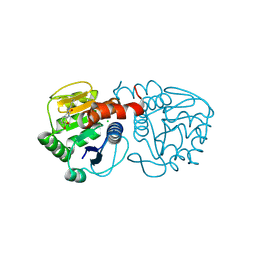 | | DJ-1 with compound 8 | | Descriptor: | CHLORIDE ION, Protein/nucleic acid deglycase DJ-1, methyl 2,3-bis(oxidanylidene)-1~{H}-indole-7-carboxylate | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFJ
 
 | | DJ-1 with compound 13 | | Descriptor: | CHLORIDE ION, Protein/nucleic acid deglycase DJ-1, butyl 2-[2,3-bis(oxidanylidene)indol-1-yl]ethanoate | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
8D5K
 
 | |
8D5E
 
 | | The complex of Gtf2b Peptide TGAASFDEF Presented by H2-Dd | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Custodio, J.M.F, Baker, B.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and physical features that distinguish tumor-controlling from inactive cancer neoepitopes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D5F
 
 | |
8D5J
 
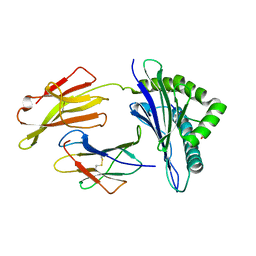 | |
1WYY
 
 | | Post-fusion hairpin conformation of the sars coronavirus spike glycoprotein | | Descriptor: | CHLORIDE ION, E2 Glycoprotein | | Authors: | Duquerroy, S, Vigouroux, A, Rottier, P.J.M, Rey, F.A, Bosch, B.J. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein
Virology, 335, 2005
|
|
3F8C
 
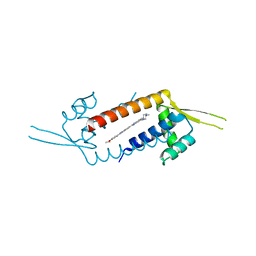 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Hoechst 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3F8B
 
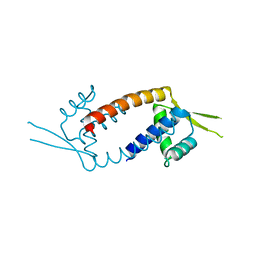 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in drug free state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
1Z3Q
 
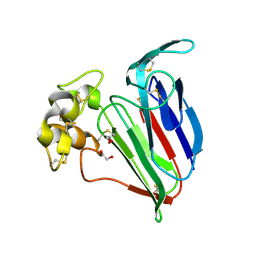 | | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7A | | Descriptor: | 1,2-ETHANEDIOL, Thaumatin-like Protein | | Authors: | Leone, P, Menu-Bouaouiche, L, Peumans, W.J, Barre, A, Payan, F, Roussel, A, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-A
Biochimie, 88, 2006
|
|
7OP0
 
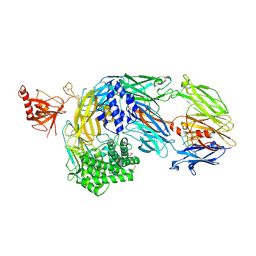 | | Crystal structure of complement C5 in complex with chemically synthesized K92 knob domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5 alpha chain, ... | | Authors: | Macpherson, A, van der Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Chemical Synthesis of Knob Domain Antibody Fragments.
Acs Chem.Biol., 16, 2021
|
|
7OU8
 
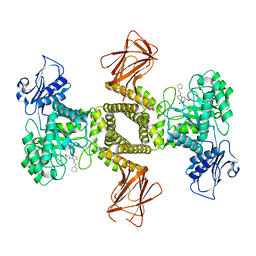 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7OU6
 
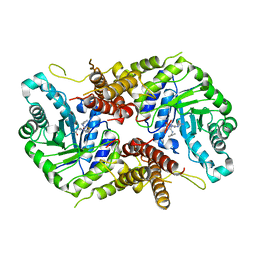 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | Protein O-GlcNAcase, ~{N}-[(3~{Z},6~{S},7~{R},8~{R},8~{a}~{S})-7,8-bis(oxidanyl)-3-(phenylmethyl)imino-1,5,6,7,8,8~{a}-hexahydro-[1,3]thiazolo[3,4-a]pyridin-6-yl]ethanamide | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7OMS
 
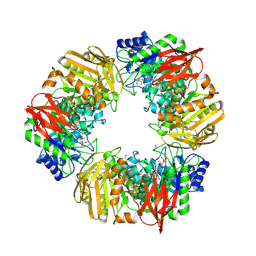 | | Bs164 in complex with mannocyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McGregor, N, Beenakker, T, Kuo, C.L, Wong, C.S, Offren, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H, Davies, G.J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
4TSO
 
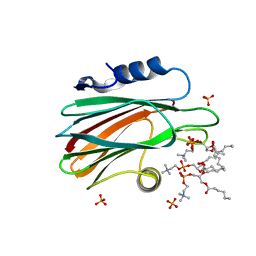 | | Crystal structure of FraC with DHPC bound (crystal form I) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSP
 
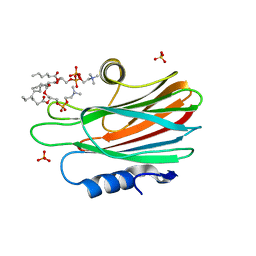 | | Crystal structure of FraC with DHPC bound (crystal form II) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSY
 
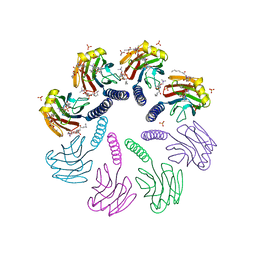 | | Crystal structure of FraC with lipids | | Descriptor: | 2-[[(E,2S,3R)-2-(hexanoylamino)-3-oxidanyl-dec-4-enoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, Fragaceatoxin C, HEPTANE-1,2,3-TRIOL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
