2GC3
 
 | |
1S0Z
 
 | | Crystal structure of the VDR LBD complexed to seocalcitol. | | Descriptor: | SEOCALCITOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
2GAK
 
 | |
1SSZ
 
 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | INFRARED SPECTROSCOPY | | Cite: | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
2IJD
 
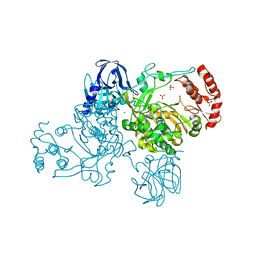 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
2IM5
 
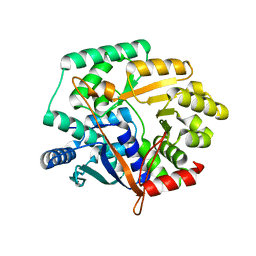 | | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas Gingivalis | | Descriptor: | Nicotinate phosphoribosyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-03 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas gingivalis
To be Published
|
|
2IML
 
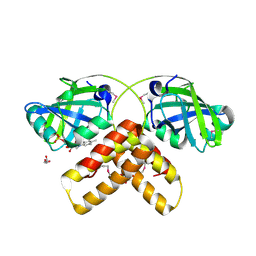 | | Crystal structure of a hypothetical protein from Archaeoglobus fulgidus binding riboflavin 5'-phosphate | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a hypothetical protein from
Archaeoglobus fulgidus binding riboflavin 5'-phosphate
To be Published
|
|
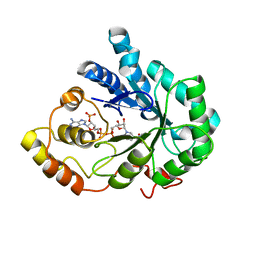
2INE
 
 | |
2IS7
 
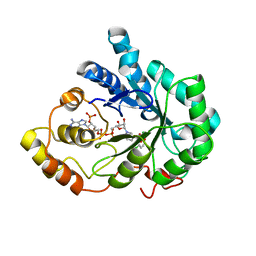 | | Crystal Structure of Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Milne, A, Brownlee, J.M. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IVI
 
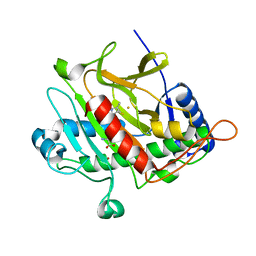 | | Isopenicillin N Synthase From Aspergillus Nidulans (Anaerobic Ac- methyl-cyclopropylglycine Fe Complex) | | Descriptor: | D-(L-A-AMINOADIPOYL)-L-CYSTEINYL-B-METHYL-D-CYCLOPROPYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Elkins, J.M, Howard-Jones, A.R, Clifton, I.J, Roach, P.L, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Interactions of Isopenicillin N Synthase with Cyclopropyl-Containing Substrate Analogues Reveal New Mechanistic Insight.
Biochemistry, 46, 2007
|
|
1T8D
 
 | | Structure of the C-type lectin domain of CD23 | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor | | Authors: | Hibbert, R.G, Teriete, P, Grundy, G.J, Sutton, B.J, Gould, H.J, McDonnell, J.M. | | Deposit date: | 2004-05-12 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of human CD23 and its interactions with IgE and CD21
J.Exp.Med., 202, 2005
|
|
2IJR
 
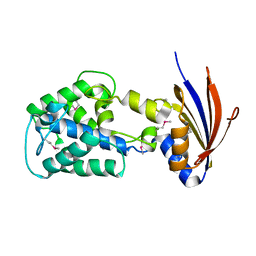 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
2ICP
 
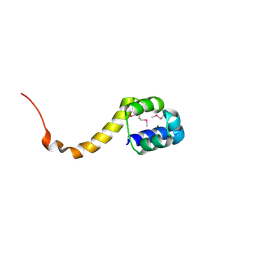 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 4.0. Northeast Structural Genomics Consortium TARGET ER390. | | Descriptor: | MAGNESIUM ION, antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the bacterial antitoxin HigA from Escherichia coli.
To be Published
|
|
2IPW
 
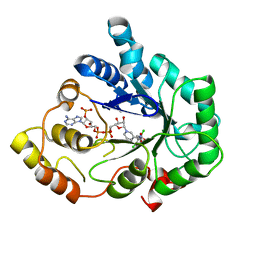 | | Crystal Structure of C298A W219Y Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IQD
 
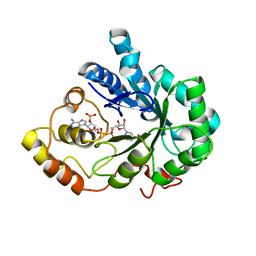 | | Crystal Structure of Aldose Reductase complexed with Lipoic Acid | | Descriptor: | Aldose reductase, LIPOIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Carlson, E, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2ICT
 
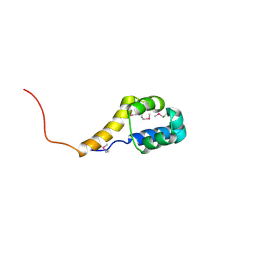 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
2ISF
 
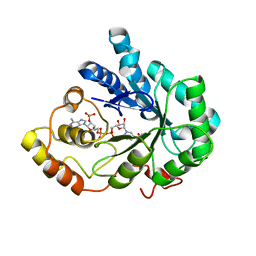 | |
1TOK
 
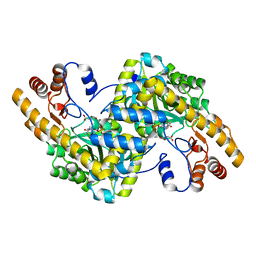 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
2IQ0
 
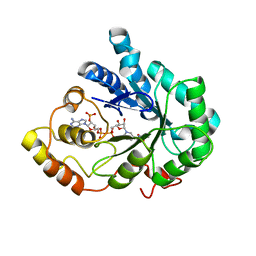 | |
1TN9
 
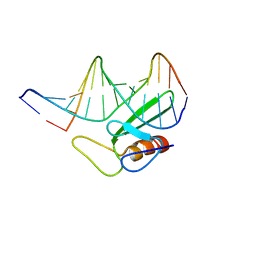 | |
1TOJ
 
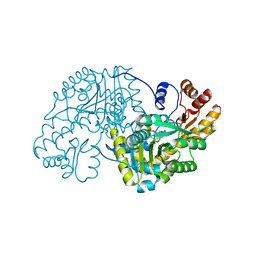 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TP9
 
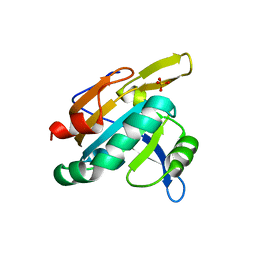 | | PRX D (type II) from Populus tremula | | Descriptor: | SULFATE ION, peroxiredoxin | | Authors: | Echalier, A, Trivelli, X, Corbier, C, Rouhier, N, Walker, O, Tsan, P, Jacquot, J.P, Krimm, I, Lancelin, J.M. | | Deposit date: | 2004-06-16 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution NMR dynamics of a D (type II) peroxiredoxin glutaredoxin and thioredoxin dependent: a new insight into the peroxiredoxin oligomerism
Biochemistry, 44, 2005
|
|
2J8P
 
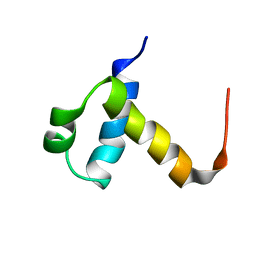 | | NMR structure of C-terminal domain of human CstF-64 | | Descriptor: | CLEAVAGE STIMULATION FACTOR 64 KDA SUBUNIT | | Authors: | Qu, X, Perez-Canadillas, J.M, Agrawal, S, De Baecke, J, Cheng, H, Varani, G, Moore, C. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domains of Vertebrate Cstf-64 and its Yeast Orthologue RNA15 Form a New Structure Critical for Mrna 3'-End Processing.
J.Biol.Chem., 282, 2007
|
|
2IDC
 
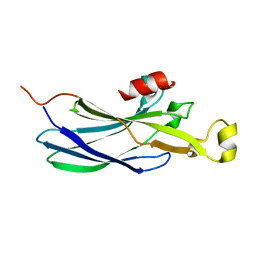 | | Structure of the Histone H3-Asf1 Chaperone Interaction | | Descriptor: | ANTI-SILENCING PROTEIN 1 AND HISTONE H3 CHIMERA | | Authors: | Antczak, A.J, Tsubota, T, Kaufman, P.D, Berger, J.M. | | Deposit date: | 2006-09-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics.
Bmc Struct.Biol., 6, 2006
|
|
2IUX
 
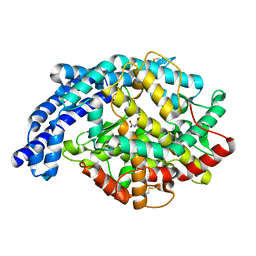 | | Human tACE mutant g1234 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Watermeyer, J.M, Swell, B.T, Natesh, R, Corradi, H.R, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-06-07 | | Release date: | 2006-10-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Testis Ace Glycosylation Mutants and Evidence for Conserved Domain Movement.
Biochemistry, 45, 2006
|
|
