4CLM
 
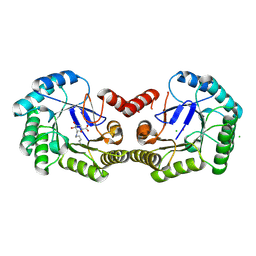 | | Structure of Salmonella typhi type I dehydroquinase irreversibly inhibited with a 1,3,4-trihydroxyciclohexane-1-carboxylic acid derivative | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Maneiro, M, Lence, E, Poza, S, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Irreversible covalent modification of type I dehydroquinase with a stable Schiff base.
Org. Biomol. Chem., 13, 2015
|
|
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
2HDA
 
 | | Yes SH3 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Yes, SULFATE ION | | Authors: | Camara-Artigas, A, Luque, I, Ruiz-Sanz, J, Mateo, P.L, Martin-Garcia, J.M. | | Deposit date: | 2006-06-20 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of the SH3 domain of the human c-Yes tyrosine kinase: Loop flexibility and amyloid aggregation.
Febs Lett., 581, 2007
|
|
1PJ1
 
 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F SOAKED WITH FERROUS IONS AT PH 5 | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PIZ
 
 | | RIBONUCLEOTIDE REDUCTASE R2 D84E MUTANT SOAKED WITH FERROUS IONS AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
2HE4
 
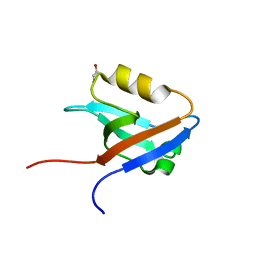 | | The crystal structure of the second PDZ domain of human NHERF-2 (SLC9A3R2) interacting with a mode 1 PDZ binding motif | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2 | | Authors: | Papagrigoriou, E, Elkins, J.M, Berridge, G, Gileady, O, Colebrook, S, Gileadi, C, Salah, E, Savitsky, P, Pantic, N, Gorrec, F, Bunkoczi, G, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
4CNP
 
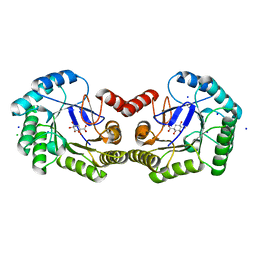 | | Structure of the Salmonella typhi type I dehydroquinase inhibited by a 3-epiquinic acid derivative | | Descriptor: | (2S)-2-HYDROXY-3-EPIQUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mechanistic Insight Into the Reaction Catalyzed by Type I Dehydroquinase
To be Published
|
|
3ISV
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with acetate ion
To be Published
|
|
1PIU
 
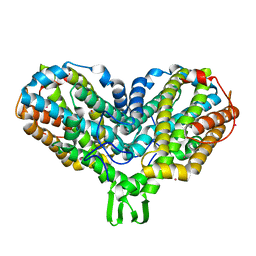 | | OXIDIZED RIBONUCLEOTIDE REDUCTASE R2-D84E MUTANT CONTAINING OXO-BRIDGED DIFERRIC CLUSTER | | Descriptor: | FE (III) ION, MERCURY (II) ION, OXYGEN ATOM, ... | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
2GZV
 
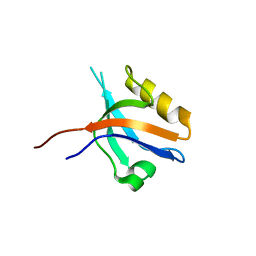 | | The cystal structure of the PDZ domain of human PICK1 | | Descriptor: | PRKCA-binding protein | | Authors: | Debreczeni, J.E, Elkins, J.M, Yang, X, Berridge, G, Bray, J, Colebrook, S, Smee, C, Savitsky, P, Gileadi, O, Turnbull, A, von Delft, F, Doyle, D.A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
3IST
 
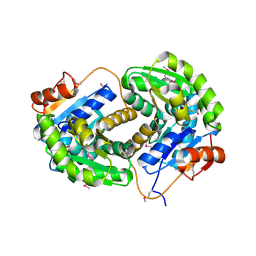 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid | | Descriptor: | CHLORIDE ION, Glutamate racemase, SUCCINIC ACID | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid
TO BE PUBLISHED
|
|
3IVR
 
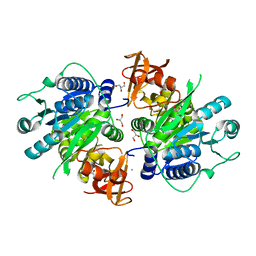 | | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA ligase FROM Rhodopseudomonas palustris CGA009 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative long-chain-fatty-acid CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA SYNTHASE FROM Rhodopseudomonas palustris CGA009
To be Published
|
|
1PQR
 
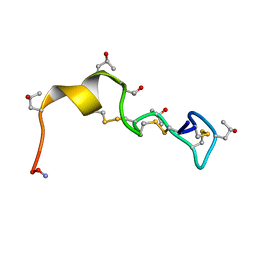 | | Solution Conformation of alphaA-Conotoxin EIVA | | Descriptor: | Alpha-A-conotoxin EIVA | | Authors: | Chi, S.-W, Park, K.-H, Suk, J.-E, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Conformation of alphaA-conotoxin EIVA, a Potent Neuromuscular Nicotinic Acetylcholine Receptor Antagonist from Conus ermineus
J.Biol.Chem., 278, 2003
|
|
1PPQ
 
 | | NMR structure of 16th module of Immune Adherence Receptor, Cr1 (Cd35) | | Descriptor: | Complement receptor type 1 | | Authors: | O'Leary, J.M, Bromek, K, Black, G.M, Uhrinova, S, Schmitz, C, Krych, M, Atkinson, J.P, Uhrin, D, Barlow, P.N. | | Deposit date: | 2003-06-17 | | Release date: | 2004-05-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces.
Protein Sci., 13, 2004
|
|
2INZ
 
 | | Crystal Structure of Aldose Reductase complexed with 2-Hydroxyphenylacetic Acid | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Carlson, E, Brownlee, J.M. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3IYC
 
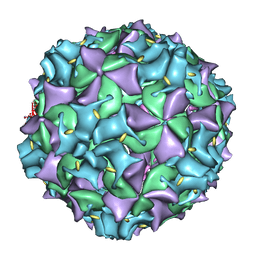 | | Poliovirus late RNA-release intermediate | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Genome polyprotein, ... | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
2IUL
 
 | | Human tACE g13 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Watermeyer, J.M, Sewell, B.T, Natesh, R, Corradi, H.R, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-06-06 | | Release date: | 2006-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Testis Ace Glycosylation Mutants and Evidence for Conserved Domain Movement.
Biochemistry, 45, 2006
|
|
2IEC
 
 | | Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri | | Descriptor: | MAGNESIUM ION, Uncharacterized protein conserved in archaea | | Authors: | Bonanno, J.B, Ramagopal, U.A, Dickey, M, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri
TO BE PUBLISHED
|
|
2J7O
 
 | | STRUCTURE OF THE RNAI POLYMERASE FROM NEUROSPORA CRASSA | | Descriptor: | MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
1RHT
 
 | | 24-MER RNA HAIRPIN COAT PROTEIN BINDING SITE FOR BACTERIOPHAGE R17 (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RNA (5'-R(P*GP*GP*GP*AP*CP*UP*GP*AP*CP*GP*AP*UP*CP*AP*CP*GP*CP*AP*GP*UP*CP*UP*AP*U)-3') | | Authors: | Borer, P.N, Lin, Y, Wang, S, Roggenbuck, M.W, Gott, J.M, Uhlenbeck, O.C, Pelczer, I. | | Deposit date: | 1995-03-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Biochemistry, 34, 1995
|
|
3J4A
 
 | | Structure of gp8 connector protein | | Descriptor: | Head-to-tail joining protein | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3KAA
 
 | | Structure of Tim-3 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Hepatitis A virus cellular receptor 2 | | Authors: | Ballesteros, A, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells.
J.Immunol., 184, 2010
|
|
2IJF
 
 | |
2IM9
 
 | | Crystal structure of protein LPG0564 from Legionella pneumophila str. Philadelphia 1, Pfam DUF1460 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Powell, A, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of hypothetical protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
TO BE PUBLISHED
|
|
