5IS9
 
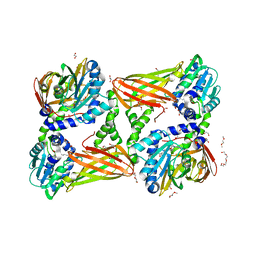 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0375 | | Descriptor: | (2S)-2-amino-4-[(3-{4-[(2S)-2-amino-2-carboxyethyl]-1H-1,2,3-triazol-1-yl}propyl){[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}amino]butanoic acid (non-preferred name), 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0375
To Be Published
|
|
5ISA
 
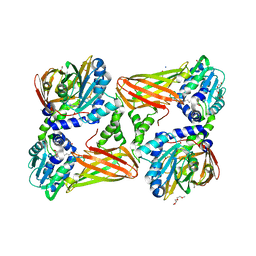 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0401 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azidopropyl)amino]-2-azanyl-butanoic acid, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0401
To Be Published
|
|
2N09
 
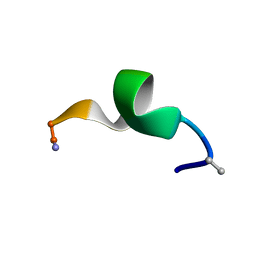 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
7KDQ
 
 | |
5IWD
 
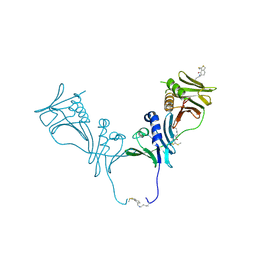 | | HCMV DNA polymerase subunit UL44 complex with a small molecule | | Descriptor: | 5-methylidene-3-(methylsulfanyl)-2-benzothiophen-4(5H)-one, DNA polymerase processivity factor | | Authors: | Chen, H, Coen, D.M, Hogle, J.M, Filman, D.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions.
ACS Infect Dis, 3, 2017
|
|
7L5M
 
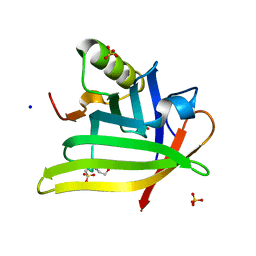 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
2ND3
 
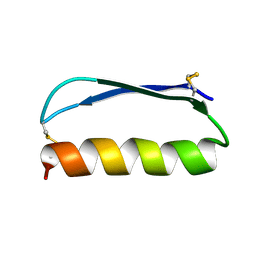 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
7L5L
 
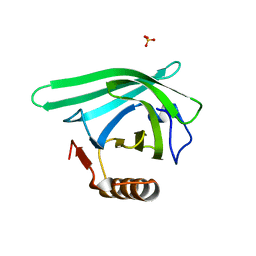 | |
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
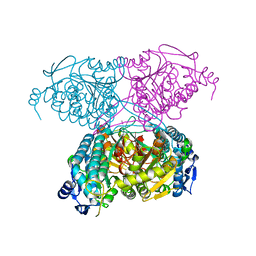
5IUU
 
 | |
2O56
 
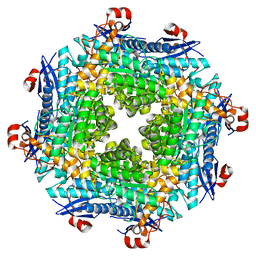 | | Crystal Structure of a Member of the Enolase Superfamily from Salmonella Typhimurium | | Descriptor: | MAGNESIUM ION, Putative mandelate racemase | | Authors: | Patskovsky, Y, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Enolase from Salmonella Typhimurium Lt2
To be Published
|
|
2O8R
 
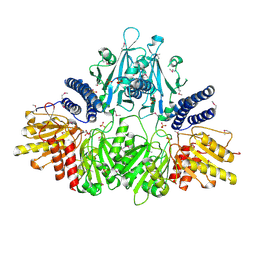 | | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis | | Descriptor: | Polyphosphate kinase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-12 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis
To be Published
|
|
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
6WLF
 
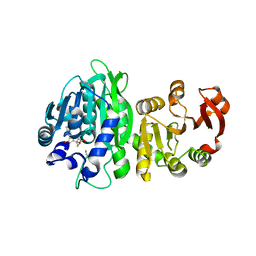 | |
6X2O
 
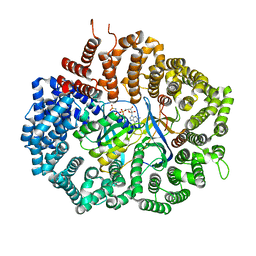 | | Crystal Structure of unliganded CRM1(E571K)-Ran-RanBP1 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2Y
 
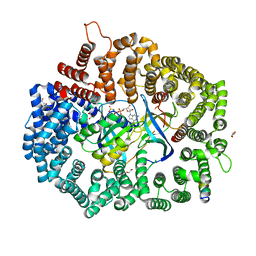 | |
6X2X
 
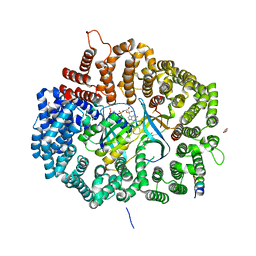 | | Crystal Structure of Mek1NES peptide bound to CRM1(E571K) | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2R
 
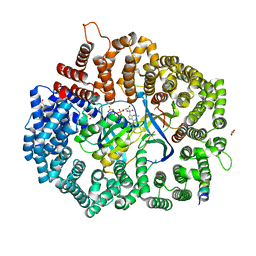 | | Crystal Structure of the 4E-TNES peptide bound to CRM1 | | Descriptor: | Eukaryotic translation initiation factor 4E transporter, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6RRH
 
 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6WFV
 
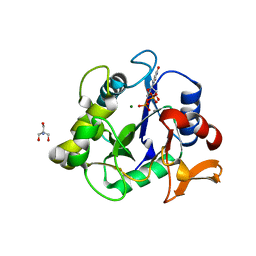 | | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Guo, H.-F, Tsai, C.-L, Miller, M.D, Phillips Jr, G.N, Tainer, J.A, Kurie, J.M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human
To Be Published
|
|
6WJL
 
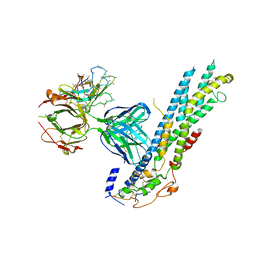 | | Crystal structure of Glypican-2 core protein in complex with D3 Fab | | Descriptor: | D3 Fab Heavy chain, D3 Fab Light Chain, Glypican-2, ... | | Authors: | Raman, S, Maris, J.M, Bosse, K.R, Julien, J.P. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A GPC2 antibody-drug conjugate is efficacious against neuroblastoma and small-cell lung cancer via binding a conformational epitope.
Cell Rep Med, 2, 2021
|
|
5MXX
 
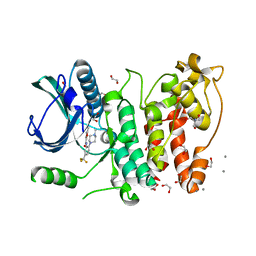 | | Crystal structure of human SR protein kinase 1 (SRPK1) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, ... | | Authors: | Tallant, C, Redondo, C, Batson, J, Toop, H.D, Babaebi-Jadidib, R, Savitsky, P, Elkins, J.M, Newman, J.A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bates, D.O, Morris, J.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
6XGR
 
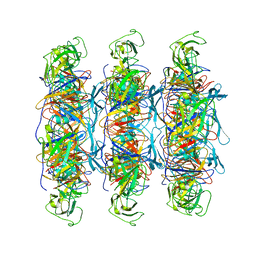 | | YSD1 major tail protein | | Descriptor: | YSD1_22 major tail protein | | Authors: | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
6X6K
 
 | | Cryo-EM Structure of the Helicobacter pylori dCag3 OMC | | Descriptor: | Cag pathogenicity island protein, Cag pathogenicity island protein (Cag7), Type IV secretion system apparatus protein CagX | | Authors: | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | Deposit date: | 2020-05-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
6X9L
 
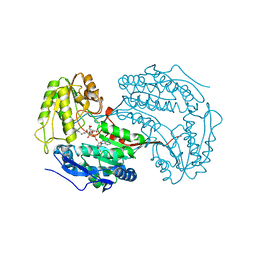 | |
