2AWN
 
 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ATP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1G4D
 
 | |
2YI9
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus in complex with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2AU1
 
 | | Crystal Structure of group A Streptococcus MAC-1 orthorhombic form | | Descriptor: | BETA-MERCAPTOETHANOL, IgG-degrading protease | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-26 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2AVO
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2Y6S
 
 | | Structure of an Ebolavirus-protective antibody in complex with its mucin-domain linear epitope | | Descriptor: | ENVELOPE GLYCOPROTEIN, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Olal, D.O, Kuehne, A, Lee, J.E, Bale, S, Dye, J.M, Saphire, E.O. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Ebola Virus-Protective Antibody in Complex with its Mucin-Domain Linear Epitope.
J.Virol., 86, 2012
|
|
2YIA
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2QR4
 
 | | Crystal structure of oligoendopeptidase-F from Enterococcus faecium | | Descriptor: | Peptidase M3B, oligoendopeptidase F | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Bain, K, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of oligoendopeptidase-F from Enterococcus faecium.
To be Published
|
|
3KQ5
 
 | | Crystal structure of an uncharacterized protein from Coxiella burnetii | | Descriptor: | Hypothetical cytosolic protein | | Authors: | Bonanno, J.B, Freeman, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-17 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Coxiella burnetii
To be Published
|
|
2Y76
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-5-ylmethoxy)-2-(benzo(b) thiophen-5-ylmethyl)-1,4,5-trihydroxycyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[b]THIOPHEN-5-YL)METHOXY-2-(BENZO[b]THIOPHEN-5-YL)METHYL-1,4,5-TRIHYDROXYCYCLOHEX-2-ENE-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2QWT
 
 | | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Adams, J, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii.
To be Published
|
|
6WW5
 
 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
2AVQ
 
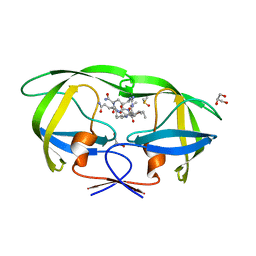 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2YKT
 
 | | Crystal structure of the I-BAR domain of IRSp53 (BAIAP2) in complex with an EHEC derived Tir peptide | | Descriptor: | BRAIN-SPECIFIC ANGIOGENESIS INHIBITOR 1-ASSOCIATED PROTEIN 2, SULFATE ION, TRANSLOCATED INTIMIN RECEPTOR PROTEIN | | Authors: | de Groot, J.C, Schlueter, K, Carius, Y, Quedenau, C, Vingadassalom, D, Faix, J, Weiss, S.M, Reichelt, J, Standfuss-Gabisch, C, Lesser, C.F, Leong, J.M, Heinz, D.W, Buessow, K, Stradal, T.E.B. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Complex Formation between Human Irsp53 and the Translocated Intimin Receptor Tir of Enterohemorrhagic E. Coli.
Structure, 19, 2011
|
|
1FXH
 
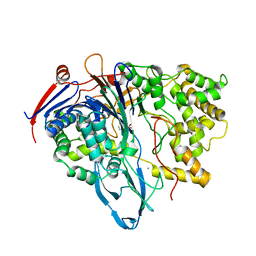 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
2YI8
 
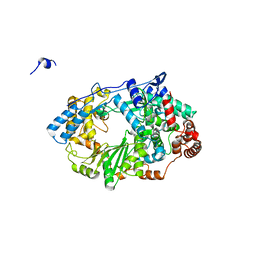 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | CHLORIDE ION, POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2V8O
 
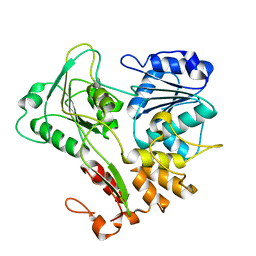 | | Structure of the Murray Valley encephalitis virus RNA helicase to 1. 9A resolution | | Descriptor: | FLAVIVIRIN PROTEASE NS3 | | Authors: | Mancini, E.J, Assenberg, R, Verma, A, Walter, T.S, Tuma, R, Grimes, J.M, Owens, R.J, Stuart, D.I. | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Murray Valley Encephalitis Virus RNA Helicase at 1.9 A Resolution.
Protein Sci., 16, 2007
|
|
6WU1
 
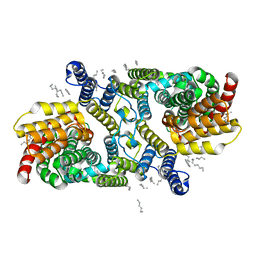 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
2CKU
 
 | |
3MOG
 
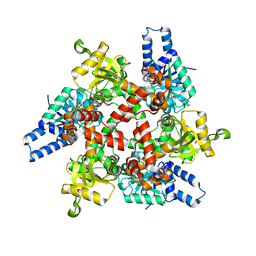 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
2PXC
 
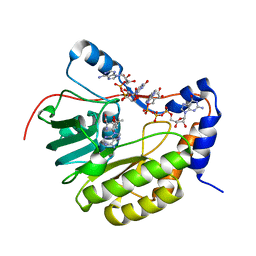 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAM and GTPA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYLMETHIONINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
3MN1
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
2PX8
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and 7M-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
3MFC
 
 | | Computationally designed end0-1,4-beta,xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3KNR
 
 | | Bacillus cereus metallo-beta-lactamase Cys221Asp mutant, 1 mM Zn(II) | | Descriptor: | ACETIC ACID, Beta-lactamase 2, GLYCEROL, ... | | Authors: | Medrano Martin, F.J, Gonzalez, J.M, Vila, A.J. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Metallo-beta-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions.
Nat.Chem.Biol., 8, 2012
|
|
