5LO9
 
 | | Thiosulfate dehydrogenase (TsdBA) from Marichromatium purpuratum - "as isolated" form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome C, ... | | Authors: | Brito, J.A, Kurth, J.M, Reuter, J, Flegler, A, Koch, T, Franke, T, Klein, E, Rowe, S, Butt, J.N, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Electron Accepting Units of the Diheme Cytochrome c TsdA, a Bifunctional Thiosulfate Dehydrogenase/Tetrathionate Reductase.
J.Biol.Chem., 291, 2016
|
|
3N2G
 
 | | TUBULIN-NSC 613863: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Barbier, P, Dorleans, A, Devred, F, Sanz, L, Allegro, D, Alfonso, C, Knossow, M, Peyrot, V, Andreu, J.M. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers.
J.Biol.Chem., 285, 2010
|
|
6QEM
 
 | | E. coli DnaBC complex bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein DnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
3N3D
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
3N6J
 
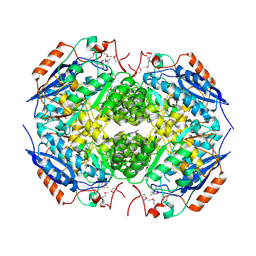 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z | | Descriptor: | Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z
To be Published
|
|
6LDV
 
 | |
6QBP
 
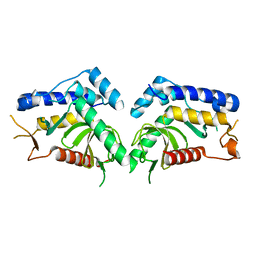 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203D | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Ramos, N, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203D
To Be Published
|
|
6QIU
 
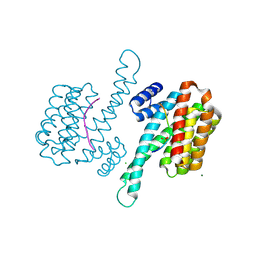 | | Crystal structure of 14-3-3 sigma in complex with Ataxin-1 Ser776 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Ataxin-1 phosphopeptide, CHLORIDE ION, ... | | Authors: | Leysen, S, Milroy, L.G, Davis, J.M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2019-01-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1
To Be Published
|
|
5MBS
 
 | |
6F5O
 
 | |
6EVS
 
 | | Characterization of 2-deoxyribosyltransferase from psychrotolerant bacterium Bacillus psychrosaccharolyticus: a suitable biocatalyst for the industrial synthesis of antiviral and antitumoral nucleosides | | Descriptor: | N-deoxyribosyltransferase | | Authors: | Fresco-Tabohada, A, Fernandez-Lucas, J, Acebal, C, Arroyo, M, Ramon, F, Mancheno, J.M, de la Mata, I. | | Deposit date: | 2017-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2'-Deoxyribosyltransferase from Bacillus psychrosaccharolyticus: A Mesophilic-Like Biocatalyst for the Synthesis of Modified Nucleosides from a Psychrotolerant Bacterium
Catalysts, 2019
|
|
3NH5
 
 | | Crystal structure of E177A-mutant murine aminoacylase 3 | | Descriptor: | ACETATE ION, Aspartoacylase-2, CHLORIDE ION, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NHI
 
 | | Crystal structure of the AnSt-D7L1-leukotriene C4 complex | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, D7 protein, GLYCEROL, ... | | Authors: | Andersen, J.F, Alvarenga, P.H, Francischetti, I.M, Calvo, E, Sa-Nunes, A, Ribeiro, J.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The function and three-dimensional structure of a thromboxane a(2)/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite.
Plos Biol., 8, 2010
|
|
8GDL
 
 | | Acid phosphatase pseudoenzyme from flea | | Descriptor: | PALMITOLEIC ACID, SEROTONIN, SULFATE ION, ... | | Authors: | Lu, S, Andersen, J.F, Ribeiro, J.M. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Acid phosphatase-like proteins, a biogenic amine and leukotriene-binding salivary protein family from the flea Xenopsylla cheopis.
Commun Biol, 6, 2023
|
|
3NHT
 
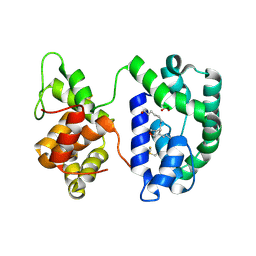 | | Crystal structure of the AnSt-D7L1-U46619 complex | | Descriptor: | (5E)-7-{6-[(1E)-3-HYDROXYOCT-1-ENYL]-2-OXABICYCLO[2.2.1]HEPT-5-YL}HEPT-5-ENOIC ACID, D7 protein, SULFATE ION | | Authors: | Andersen, J.F, Alvarenga, P.H, Francischetti, I.M, Calvo, E, Sa-Nunes, A, Ribeiro, J.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The function and three-dimensional structure of a thromboxane a(2)/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite.
Plos Biol., 8, 2010
|
|
8FYZ
 
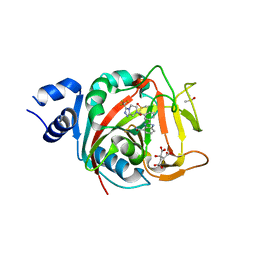 | | Crystal structure of human PARP1 ART domain bound to inhibitor UKTT10 (compound 13) | | Descriptor: | (2P)-2-{3-[(4R)-3-(trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazine-7(8H)-carbonyl]phenyl}-1H-benzimidazole-4-carboxamide, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks.
Biochem.J., 481, 2024
|
|
8FYY
 
 | |
8G0H
 
 | |
8FZ1
 
 | | Crystal structure of human PARP1 ART domain bound to inhibitor UKTT22 (compound 14) | | Descriptor: | (2P)-2-{3-[(2-amino-4,5-dimethylphenyl)carbamoyl]phenyl}-1H-benzimidazole-4-carboxamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks.
Biochem.J., 481, 2024
|
|
5MPA
 
 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Q1L
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
5MRD
 
 | | Human PDK1-PKCiota Kinase Chimera in Complex with Allosteric Compound PS267 Bound to the PIF-Pocket | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Arencibia, J.M, Froehner, W, Krupa, M, Pastor-Flores, D, Merker, P, Oellerich, T, Neimanis, S, Schmithals, C, Koeberle, V, Suess, E, Zeuzem, S, Stark, H, Piiper, A, Odadzic, D, Schulze, J.O, Biondi, R.M. | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | An Allosteric Inhibitor Scaffold Targeting the PIF-Pocket of Atypical Protein Kinase C Isoforms.
ACS Chem. Biol., 12, 2017
|
|
5MOI
 
 | | Crystal structure of human IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
6EYO
 
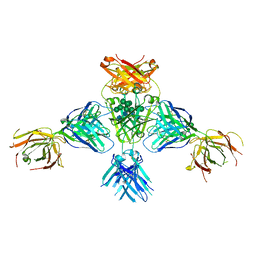 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6F64
 
 | | Crystal structure of the SYCP1 C-terminal back-to-back assembly | | Descriptor: | ACETATE ION, Synaptonemal complex protein 1 | | Authors: | Dunce, J.M, Millan, C, Uson, I, Davies, O.R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-06-06 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
