2KV9
 
 | | Integrin beta3 subunit in a disulfide linked alphaIIb-beta3 cytosolic domain | | Descriptor: | Integrin beta-3 | | Authors: | Metcalf, D.G, Kielec, J.M, Valentine, K.G, Wand, A, Bennett, J.S, William, D.F, Moore, D.T, Molnar, K. | | Deposit date: | 2010-03-10 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the {alpha}IIb{beta}3 cytoplasmic interaction suggests a mechanism for integrin regulation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KWB
 
 | | Minimal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from C.elegans, Northeast Structural Genomics Consortium Target WR73 | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Aramini, J.M, Rossi, P, Cort, J.R, Cooper, B, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Minimal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from C.elegans, Northeast Structural Genomics Consortium Target WR73
To be Published
|
|
2K5D
 
 | | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108]. | | Descriptor: | uncharacterized protein SAG0934 | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Foote, E.L, Jiang, M, Xiao, R, Sharma, S, Swapna, G.VT, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108].
To be Published
|
|
2KW6
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 1 (CDK2-associated protein 1; oral cancer suppressor Deleted in oral cancer 1, DOC-1) from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3057H | | Descriptor: | Cyclin-dependent kinase 2-associated protein 1 | | Authors: | Ertekin, A, Aramini, J.M, Rossi, P, Lee, A.B, Jiang, M, Ciccosanti, C.T, Xiao, R, Swapna, G.V.T, Rost, B, Everett, J.K, Acton, T.B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Human cyclin-dependent kinase 2-associated protein 1 (CDK2AP1) is dimeric in its disulfide-reduced state, with natively disordered N-terminal region.
J.Biol.Chem., 287, 2012
|
|
2L7M
 
 | |
2L9Z
 
 | | Zinc knuckle in PRDM4 | | Descriptor: | PR domain zinc finger protein 4, ZINC ION | | Authors: | Briknarova, K, Atwater, D.Z, Glicken, J.M, Maynard, S.J, Ness, T.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The PR/SET domain in PRDM4 is preceded by a zinc knuckle.
Proteins, 79, 2011
|
|
1UVM
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5NT RNA conformation A | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UO4
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, iodobenzene | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1VZ8
 
 | |
5CIN
 
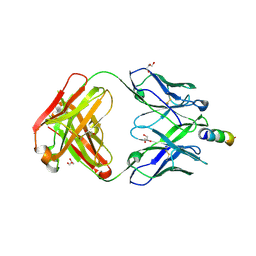 | | Crystal Structure of non-neutralizing version of 4E10 (DeltaLoop) with epitope bound | | Descriptor: | CHLORIDE ION, FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
1WO3
 
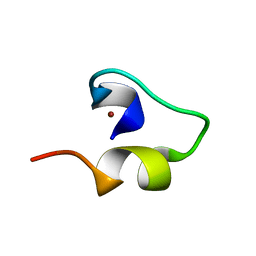 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | Descriptor: | RNA | | Authors: | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 2002-07-17 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
1X3Z
 
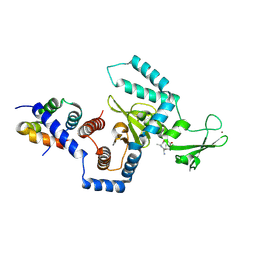 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4WV9
 
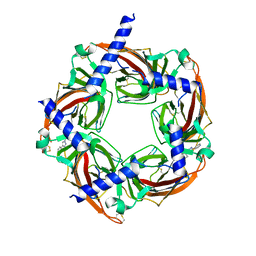 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane | | Descriptor: | (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J.M, Sankaran, B. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound.
To Be Published
|
|
1WO6
 
 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WPD
 
 | | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins | | Descriptor: | Potassium channel toxin alpha-KTx 6.2,Potassium channel toxin alpha-KTx 6.3 | | Authors: | Regaya, I, Beeton, C, Ferrat, G, Andreotti, N, Chandy, G.K, Darbon, H, De Waard, M, Sabatier, J.M. | | Deposit date: | 2004-09-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins
J.Biol.Chem., 279, 2004
|
|
2L7F
 
 | |
1WL6
 
 | | Mg-substituted form of E. coli aminopeptidase P | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-06-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional implications of metal ion selection in aminopeptidase p, a metalloprotease with a dinuclear metal center
Biochemistry, 44, 2005
|
|
1WO4
 
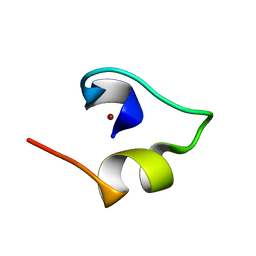 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
2LCO
 
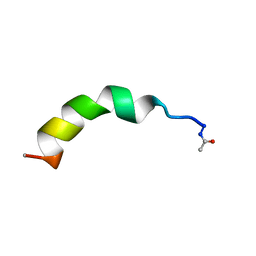 | |
2LCN
 
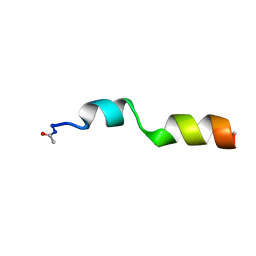 | |
1WPA
 
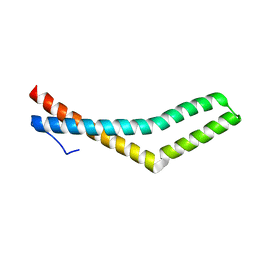 | |
2LZK
 
 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
1X3W
 
 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
