8V3M
 
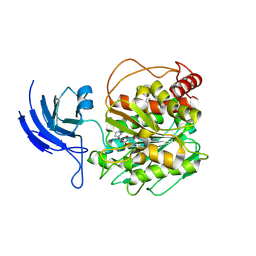 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3Q
 
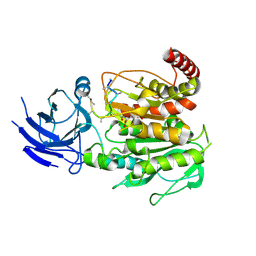 | | Structure of CCP5 class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3O
 
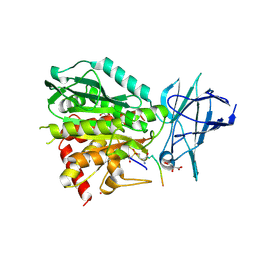 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3R
 
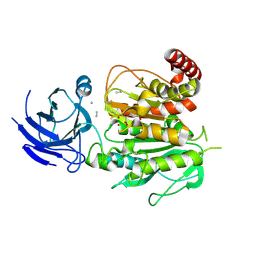 | | Structure of CCP5 class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
5BVP
 
 | |
5CB6
 
 | | Structure of adenosine-5'-phosphosulfate kinase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Herrmann, J, Jez, J.M. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Recapitulating the Structural Evolution of Redox Regulation in Adenosine 5'-Phosphosulfate Kinase from Cyanobacteria to Plants.
J.Biol.Chem., 290, 2015
|
|
5CB8
 
 | |
5CLM
 
 | | 1,4-Oxazine BACE1 inhibitors | | Descriptor: | Beta-secretase 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rombouts, F, Tresadern, G, Delgado, O, Martinez Lamenca, C, Van Gool, M, Garcia-Molina, A, Alonso De Diego, S, Oehlrich, D, Prokopcova, H, Alonso, J.M, Austin, N, Borghys, H, Van Brandt, S, Surkyn, M, De Cleyn, M, Vos, A, Alexander, R, MacDonald, G, Moechars, D, Trabanco, A, Gijsen, H. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-30 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 1,4-Oxazine beta-Secretase 1 (BACE1) Inhibitors: From Hit Generation to Orally Bioavailable Brain Penetrant Leads.
J.Med.Chem., 58, 2015
|
|
8VCI
 
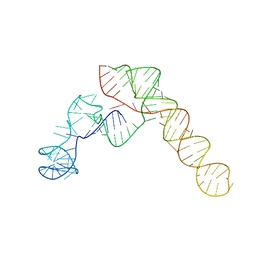 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | Descriptor: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | Authors: | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Biochemistry, 63, 2024
|
|
5MXX
 
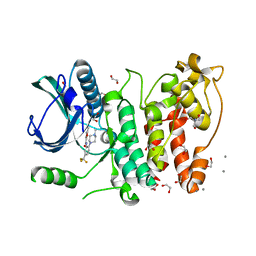 | | Crystal structure of human SR protein kinase 1 (SRPK1) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, ... | | Authors: | Tallant, C, Redondo, C, Batson, J, Toop, H.D, Babaebi-Jadidib, R, Savitsky, P, Elkins, J.M, Newman, J.A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bates, D.O, Morris, J.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
6XZP
 
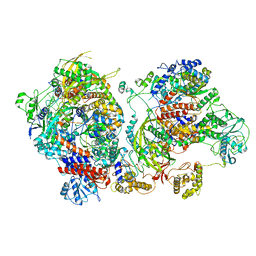 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 4 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
5CFP
 
 | | Crystal structure of anemone STING (Nematostella vectensis) 'humanized' F276K in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p]' | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
8AN1
 
 | |
6YBP
 
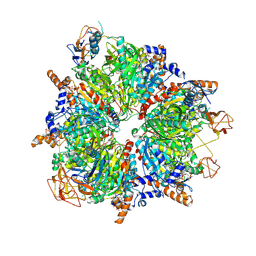 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
5NKS
 
 | | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4) | | Descriptor: | Dihydropyrimidinase-related protein 4 | | Authors: | Mathea, S, Elkins, J.M, Strain-Damerell, C, Salah, E, Borkowska, O, Chalk, R, Burgess-Brown, N, Pinkas, D.M, von Delft, F, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4)
To Be Published
|
|
5CFO
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'rotated' open conformation | | Descriptor: | Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
6Y0C
 
 | | Influenza C virus polymerase in complex with human ANP32A - Subclass 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*GP*CP*CP*UP*UP*UP*U)-3'), ... | | Authors: | Fan, H, Carrique, L, Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-07 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
5CFL
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p] | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CITRATE ANION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
8V8Z
 
 | |
5CFR
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'unrotated' closed conformation | | Descriptor: | CALCIUM ION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5N6P
 
 | | AMPA receptor NTD mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Rossmann, M, Krieger, J.M. | | Deposit date: | 2017-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric communication pathways in glutamate receptor N-terminal domains
To Be Published
|
|
8VON
 
 | |
5CFQ
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 2',3' cGAMP, c[G(2',5')pA(3',5')p] | | Descriptor: | SULFATE ION, Stimulator of Interferon Genes, cGAMP | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
8VOM
 
 | |
6YRS
 
 | | Structure of a new variant of GNCA ancestral beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M, Modi, T, Ozkan, S.B. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hinge-shift mechanism as a protein design principle for the evolution of beta-lactamases from substrate promiscuity to specificity.
Nat Commun, 12, 2021
|
|
