1SUU
 
 | |
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
1SX0
 
 | | Solution NMR Structure and X-Ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase | | Descriptor: | SecA | | Authors: | Dempsey, B.R, Wrona, M, Moulin, J.M, Gloor, G.B, Jalilehvand, F, Lajoie, G, Shaw, G.S, Shilton, B.H. | | Deposit date: | 2004-03-30 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase.
Biochemistry, 43, 2004
|
|
3D2Y
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3OOQ
 
 | | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8 | | Descriptor: | GLYCEROL, amidohydrolase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8
To be Published
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4CIX
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with(1R,4R,5R)-1,4,5-trihydroxy-3-((1S)-1-hydroxy-2-phenyl) ethylcyclohex-2-en-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[(1S)-1-hydroxy-2-phenyl]ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
3HI2
 
 | | Structure of the N-terminal domain of the E. coli antitoxin MqsA (YgiT/b3021) in complex with the E. coli toxin MqsR (YgiU/b3022) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator mqsA(ygiT), Motility quorum-sensing regulator mqsR, ... | | Authors: | Grigoriu, S, Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
3HIG
 
 | |
1T3J
 
 | | Mitofusin domain HR2 V686M/I708M mutant | | Descriptor: | mitofusin 1 | | Authors: | Koshiba, T, Detmer, S.A, Kaiser, J.T, Chen, H, McCaffery, J.M, Chan, D.C. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of mitochondrial tethering by mitofusin complexes
Science, 305, 2004
|
|
2I52
 
 | | Crystal structure of protein PTO0218 from Picrophilus torridus, Pfam DUF372 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramagopal, U.A, Gilmore, J, Toro, R, Bain, K.T, McKenzie, C, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-23 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of hypothetical protein PTO0218 from Picrophilus torridus
To be Published
|
|
1O72
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with phosphorylcholine | | Descriptor: | PHOSPHOCHOLINE, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
3CZ5
 
 | | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1 | | Descriptor: | PHOSPHATE ION, Two-component response regulator, LuxR family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1.
To be Published
|
|
4BZZ
 
 | | Complete crystal structure of carboxylesterase Cest-2923 from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, LIPASE/ESTERASE, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
4C08
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with CaCl2 at 1.34 Angstroms | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.338 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
1OIN
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
3OX4
 
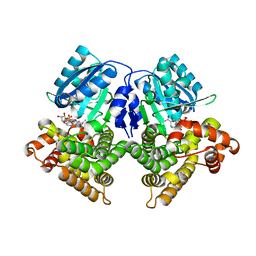 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
1Q2W
 
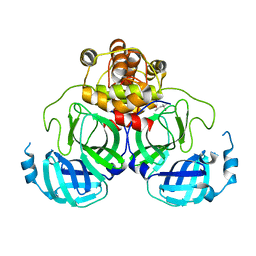 | | X-Ray Crystal Structure of the SARS Coronavirus Main Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like protease | | Authors: | Bonanno, J.B, Fowler, R, Gupta, S, Hendle, J, Lorimer, D, Romero, R, Sauder, J.M, Wei, C.L, Liu, E.T, Burley, S.K, Harris, T. | | Deposit date: | 2003-07-26 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Company Says It Mapped Part of SARS Virus
New York Times, 30 July, 2003
|
|
3F5Q
 
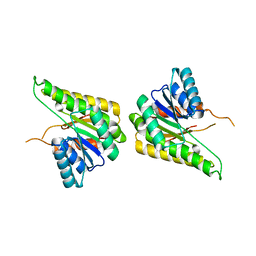 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
2FD3
 
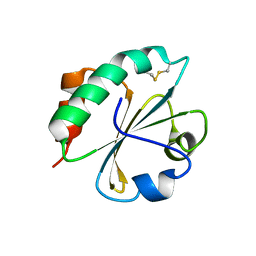 | |
2FGI
 
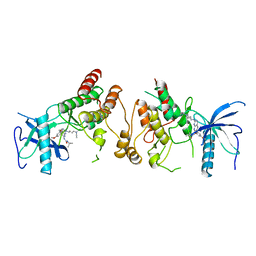 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF FGF RECEPTOR 1 IN COMPLEX WITH INHIBITOR PD173074 | | Descriptor: | 1-TERT-BUTYL-3-[6-(3,5-DIMETHOXY-PHENYL)-2-(4-DIETHYLAMINO-BUTYLAMINO)-PYRIDO[2,3-D]PYRIMIDIN-7-YL]-UREA, PROTEIN (FIBROBLAST GROWTH FACTOR (FGF) RECEPTOR 1) | | Authors: | Mohammadi, M, Froum, S, Hamby, J.M, Schroeder, M, Panek, R.L, Lu, G.H, Eliseenkova, A.V, Green, D, Schlessinger, J, Hubbard, S.R. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain.
EMBO J., 17, 1998
|
|
1OG6
 
 | |
4C06
 
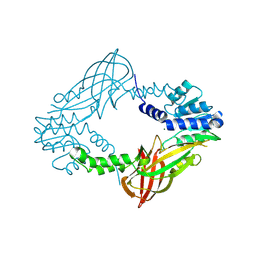 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with MgCl2 | | Descriptor: | MAGNESIUM ION, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
2FH7
 
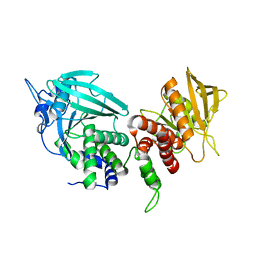 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1OJQ
 
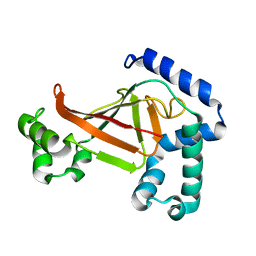 | | The crystal structure of C3stau2 from S. aureus | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-15 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
