1O2A
 
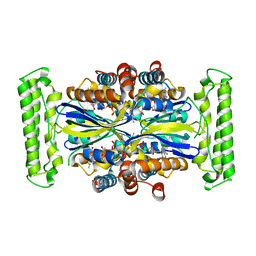 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD at 1.8 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
3JW5
 
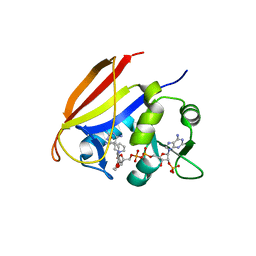 | |
3CF1
 
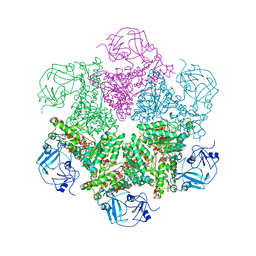 | | Structure of P97/vcp in complex with ADP/ADP.alfx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
2FWV
 
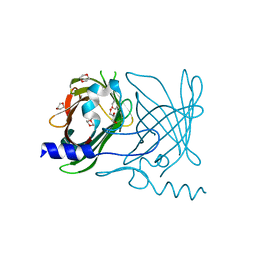 | | Crystal Structure of Rv0813 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, hypothetical protein MtubF_01000852 | | Authors: | Shepard, W, Haouz, A, Grana, M, Buschiazzo, A, Betton, J.M, Cole, S.T, Alzari, P.M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-03 | | Release date: | 2006-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Rv0813c from Mycobacterium tuberculosis Reveals a New Family of Fatty Acid-Binding Protein-Like Proteins in Bacteria
J.Bacteriol., 189, 2007
|
|
3HM7
 
 | | Crystal structure of allantoinase from Bacillus halodurans C-125 | | Descriptor: | Allantoinase, ZINC ION | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Miller, S, Wasserman, S.R, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Allantoinase from Bacillus Halodurans
To be Published
|
|
1M9O
 
 | |
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
2FZ6
 
 | | Crystal structure of hydrophobin HFBI | | Descriptor: | Hydrophobin-1, ZINC ION | | Authors: | Hakanpaa, J.M, Rouvinen, J. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of Trichoderma reesei hydrophobin HFBI--The structure of a protein amphiphile with and without detergent interaction.
Protein Sci., 15, 2006
|
|
1SM1
 
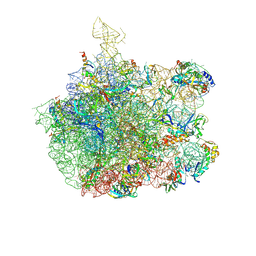 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH QUINUPRISTIN AND DALFOPRISTIN | | Descriptor: | 23S RIBOSOMAL RNA, 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Harms, J.M, Schluenzen, F, Fucini, P, Bartels, H, Yonath, A. | | Deposit date: | 2004-03-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Alterations at the Peptidyl Transferase Centre of the Ribosome Induced by the Synergistic Action of the Streptogramins Dalfopristin and Quinupristin.
Bmc Biol., 2, 2004
|
|
4B88
 
 | |
1S75
 
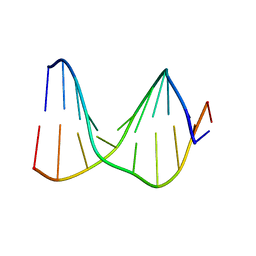 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
1S84
 
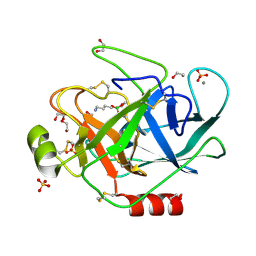 | | PORCINE TRYPSIN COVALENT COMPLEX WITH 4-AMINO BUTANOL, BORATE AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3,2-DIOXABOROLAN-2-YLOXY)BUTAN-1-AMINIUM, CALCIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
4BKJ
 
 | | Crystal structure of the human DDR1 kinase domain in complex with imatinib | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Mechanisms Determining Inhibition of the Collagen Receptor Ddr1 by Selective and Multi-Targeted Type II Kinase Inhibitors
J.Mol.Biol., 426, 2014
|
|
2FMP
 
 | | DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
3QQT
 
 | |
3JWF
 
 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and (R)-2,4-diamino-5-(3-hydroxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP113A) | | Descriptor: | (1R)-3-(2,4-diamino-6-methylpyrimidin-5-yl)-1-(3,4,5-trimethoxyphenyl)prop-2-yn-1-ol, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
4AW1
 
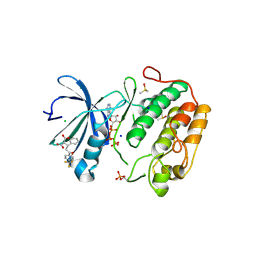 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS210 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
3K9D
 
 | | CRYSTAL STRUCTURE OF PROBABLE ALDEHYDE DEHYDROGENASE FROM Listeria monocytogenes EGD-e | | Descriptor: | ALDEHYDE DEHYDROGENASE, CHLORIDE ION, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase from Listeria Monocytogenes
To be Published
|
|
1M1J
 
 | | Crystal structure of native chicken fibrinogen with two different bound ligands | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, Z, Kollman, J.M, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Native Chicken Fibrinogen at 2.7 A Resolution
Biochemistry, 40, 2001
|
|
4BBB
 
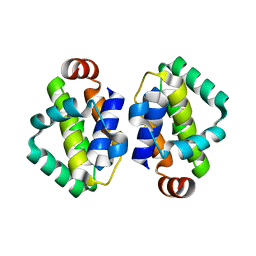 | | THE STRUCTURE OF VACCINIA VIRUS N1 Q61Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
3KAK
 
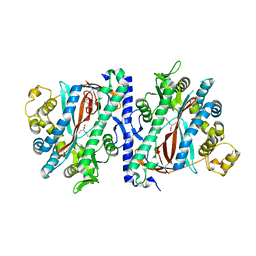 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3I15
 
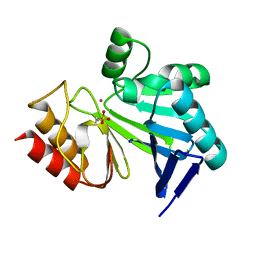 | |
3J2P
 
 | | CryoEM structure of Dengue virus envelope protein heterotetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
1M7T
 
 | |
8CYO
 
 | | Nurr1 Covalently Bound to a Synthetic Ligand, 10.25, via a Disulfide Bond | | Descriptor: | 2-(3,4-dichlorophenoxy)-N-(2-sulfanylethyl)acetamide, CHLORIDE ION, Nuclear receptor subfamily 4 group A member 2 | | Authors: | Bruning, J.M, Liu, J, England, P.M. | | Deposit date: | 2022-05-24 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Nurr1 Covalently Bound to a Synthetic Ligand, 10.25, via a Disulfide Bond
To Be Published
|
|
