1L8Q
 
 | |
4CS8
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 2 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
2FO0
 
 | | Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Seeliger, M, Davies, J.M, Weis, W.I, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase.
Mol.Cell, 21, 2006
|
|
3MJM
 
 | | His257Ala mutant of dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Ernberg, K.E, Guss, J.M, Lee, M, Maher, M.J. | | Deposit date: | 2010-04-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | His257Ala mutant of dihydroorotase from E. coli
To be Published
|
|
1PV4
 
 | |
4CI6
 
 | | Mechanisms of crippling actin-dependent phagocytosis by YopO | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Lee, W.L, Grimes, J.M, Robinson, R.C. | | Deposit date: | 2013-12-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Yersinia Effector Yopo Uses Actin as Bait to Phosphorylate Proteins that Regulate Actin Polymerization.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1PVO
 
 | |
4CS7
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 1 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
3FZV
 
 | | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator, SULFATE ION | | Authors: | Knapik, A.A, Tkaczuk, K.L, Chruszcz, M, Wang, S, Zimmerman, M.D, Cymborowski, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Bujnicki, J.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
3DBI
 
 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
2FQQ
 
 | | Crystal structure of human caspase-1 (Cys285->Ala, Cys362->Ala, Cys364->Ala, Cys397->Ala) in complex with 1-methyl-3-trifluoromethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid (2-mercapto-ethyl)-amide | | Descriptor: | 1-METHYL-3-TRIFLUOROMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID (2-MERCAPTO-ETHYL)-AMIDE, Caspase-1 | | Authors: | Scheer, J.M, Wells, J.A, Romanowski, M.J. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A common allosteric site and mechanism in caspases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3M2M
 
 | |
3DGP
 
 | | Crystal Structure of the complex between Tfb5 and the C-terminal domain of Tfb2 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, RNA polymerase II transcription factor B subunit 5 | | Authors: | Kainov, D.E, Cavarelli, J, Egly, J.M, Poterszman, A. | | Deposit date: | 2008-06-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for group A trichothiodystrophy
Nat.Struct.Mol.Biol., 15, 2008
|
|
1S0Z
 
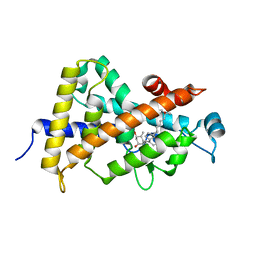 | | Crystal structure of the VDR LBD complexed to seocalcitol. | | Descriptor: | SEOCALCITOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
3FXG
 
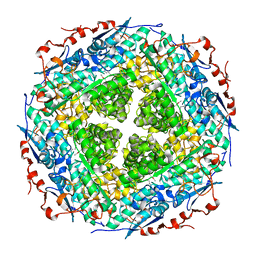 | | Crystal structure of rhamnonate dehydratase from Gibberella zeae complexed with Mg | | Descriptor: | MAGNESIUM ION, Rhamnonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rhamnonate dehydratase from Gibberella zeae complexed with Mg
To be Published
|
|
4CS9
 
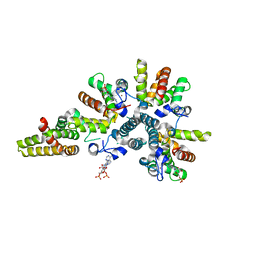 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
1S1Q
 
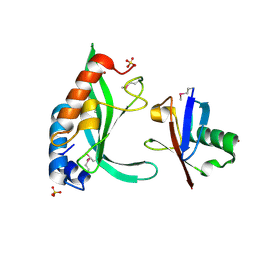 | | TSG101(UEV) domain in complex with Ubiquitin | | Descriptor: | ACETIC ACID, COPPER (II) ION, SULFATE ION, ... | | Authors: | Sundquist, W.I, Schubert, H.L, Kelly, B.N, Hill, G.C, Holton, J.M, Hill, C.P. | | Deposit date: | 2004-01-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin recognition by the human TSG101 protein
Mol.Cell, 13, 2004
|
|
1MM3
 
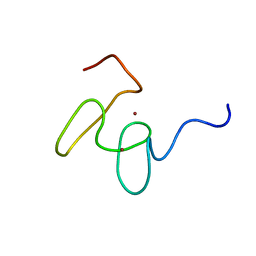 | | Solution structure of the 2nd PHD domain from Mi2b with C-terminal loop replaced by corresponding loop from WSTF | | Descriptor: | Mi2-beta(Chromodomain helicase-DNA-binding protein 4) and transcription factor WSTF, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
4BIJ
 
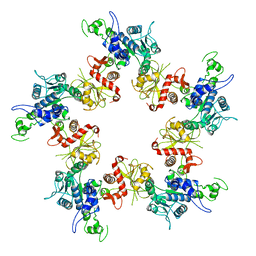 | | Threading model of T7 large terminase | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
3K4H
 
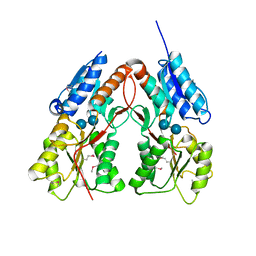 | | CRYSTAL STRUCTURE OF putative transcriptional regulator LacI from Bacillus cereus subsp. cytotoxis NVH 391-98 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, putative transcriptional regulator | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-10 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative transcriptional regulator LacI from Bacillus cereus subsp. cytotoxis NVH 391-98
To be Published
|
|
1S83
 
 | | PORCINE TRYPSIN COMPLEXED WITH 4-AMINO PROPANOL | | Descriptor: | 4-HYDROXYBUTAN-1-AMINIUM, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
3HVD
 
 | | The Protective Antigen Component of Anthrax Toxin Forms Functional Octameric Complexes | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Kintzer, A.F, Dong, K.C, Berger, J.M, Krantz, B.A. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The protective antigen component of anthrax toxin forms functional octameric complexes.
J.Mol.Biol., 392, 2009
|
|
3HY9
 
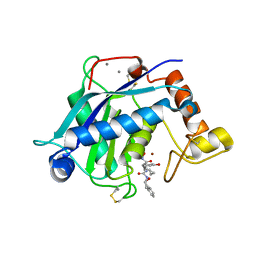 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (3R)-N~2~-(cyclopropylmethyl)-N~1~-hydroxy-3-(3-hydroxybenzyl)-N~4~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-L-aspartamide, CALCIUM ION, Catalytic Domain of ADAMTS-5, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
8ETR
 
 | | CryoEM Structure of NLRP3 NACHT domain in complex with G2394 | | Descriptor: | (6S,8R)-N-[(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)carbamoyl]-6-(methylamino)-6,7-dihydro-5H-pyrazolo[5,1-b][1,3]oxazine-3-sulfonamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Murray, J.M, Johnson, M.C. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Overcoming Preclinical Safety Obstacles to Discover ( S )- N -((1,2,3,5,6,7-Hexahydro- s -indacen-4-yl)carbamoyl)-6-(methylamino)-6,7-dihydro-5 H -pyrazolo[5,1- b ][1,3]oxazine-3-sulfonamide (GDC-2394): A Potent and Selective NLRP3 Inhibitor.
J.Med.Chem., 65, 2022
|
|
1M0I
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site | | Descriptor: | SULFATE ION, endodeoxyribonuclease I | | Authors: | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | Deposit date: | 2002-06-13 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I
Embo J., 21, 2002
|
|
